The HPSEVERITY Procedure
- Overview
-
Getting Started

-
Syntax

-
Details
 Predefined DistributionsCensoring and TruncationParameter Estimation MethodParameter InitializationEstimating Regression EffectsLevelization of Classification VariablesSpecification and Parameterization of Model EffectsEmpirical Distribution Function Estimation MethodsStatistics of FitDistributed and Multithreaded ComputationDefining a Severity Distribution Model with the FCMP ProcedurePredefined Utility FunctionsScoring FunctionsCustom Objective FunctionsInput Data SetsOutput Data SetsDisplayed OutputODS Graphics
Predefined DistributionsCensoring and TruncationParameter Estimation MethodParameter InitializationEstimating Regression EffectsLevelization of Classification VariablesSpecification and Parameterization of Model EffectsEmpirical Distribution Function Estimation MethodsStatistics of FitDistributed and Multithreaded ComputationDefining a Severity Distribution Model with the FCMP ProcedurePredefined Utility FunctionsScoring FunctionsCustom Objective FunctionsInput Data SetsOutput Data SetsDisplayed OutputODS Graphics -
Examples
 Defining a Model for Gaussian DistributionDefining a Model for the Gaussian Distribution with a Scale ParameterDefining a Model for Mixed-Tail DistributionsFitting a Scaled Tweedie Model with RegressorsFitting Distributions to Interval-Censored DataBenefits of Distributed and Multithreaded ComputingEstimating Parameters Using Cramér-von Mises EstimatorDefining a Finite Mixture Model That Has a Scale ParameterPredicting Mean and Value-at-Risk by Using Scoring FunctionsScale Regression with Rich Regression Effects
Defining a Model for Gaussian DistributionDefining a Model for the Gaussian Distribution with a Scale ParameterDefining a Model for Mixed-Tail DistributionsFitting a Scaled Tweedie Model with RegressorsFitting Distributions to Interval-Censored DataBenefits of Distributed and Multithreaded ComputingEstimating Parameters Using Cramér-von Mises EstimatorDefining a Finite Mixture Model That Has a Scale ParameterPredicting Mean and Value-at-Risk by Using Scoring FunctionsScale Regression with Rich Regression Effects - References
A finite mixture model is a stochastic model that postulates that the probability distribution of the data generation process is a mixture of a finite number of probability distributions. For example, when an insurance company analyzes loss data from multiple policies that are underwritten in different geographic regions, some regions might behave similarly, but the distribution that governs some regions might be different from the distribution that governs other regions. Further, it might not be known which regions behave similarly. Also, the larger amounts of losses might follow a different stochastic process from the stochastic process that governs the smaller amounts of losses. It helps to model all policies together in order to pool the data together and exploit any commonalities among the regions, and the use of a finite mixture model can help capture the differences in distributions across regions and ranges of loss amounts.
Formally, if ![]() and
and ![]() denote the PDF and CDF, respectively, of component distribution i and
denote the PDF and CDF, respectively, of component distribution i and ![]() represents the mixing probability that is associated with component i, then the PDF and CDF of the finite mixture of K distribution components are
represents the mixing probability that is associated with component i, then the PDF and CDF of the finite mixture of K distribution components are
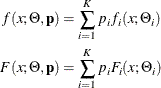
where ![]() denotes the parameters of component distribution i and
denotes the parameters of component distribution i and ![]() denotes the parameters of the mixture distribution, which is a union of all the
denotes the parameters of the mixture distribution, which is a union of all the ![]() parameters.
parameters. ![]() denotes the set of mixing probabilities. All mixing probabilities must add up to 1 (
denotes the set of mixing probabilities. All mixing probabilities must add up to 1 (![]() ).
).
You can define the finite mixture of a specific number of components and specific distributions for each of the components by defining the FCMP functions for the PDF and CDF. However, in general, it is not possible to fit a scale regression model by using any finite mixture distribution unless you take special care to ensure that the mixture distribution has a scale parameter. This example provides a formulation of a two-component finite mixture model that has a scale parameter.
To start with, each component distribution must have either a scale parameter or a log-transformed scale parameter. Let ![]() and
and ![]() denote the scale parameters of the first and second components, respectively. Let
denote the scale parameters of the first and second components, respectively. Let ![]() be the mixing probability, which makes
be the mixing probability, which makes ![]() by using the constraint on
by using the constraint on ![]() . The PDF of the mixture of these two distributions can be written as
. The PDF of the mixture of these two distributions can be written as
where ![]() and
and ![]() denote the sets of nonscale parameters of the first and second components, respectively, and
denote the sets of nonscale parameters of the first and second components, respectively, and ![]() denotes a union of
denotes a union of ![]() and
and ![]() . For the mixture to have the scale parameter
. For the mixture to have the scale parameter ![]() , the PDF must be of the form
, the PDF must be of the form
where ![]() ,
, ![]() , and
, and ![]() denote the modified sets of nonscale parameters. One simple way to achieve this is to make
denote the modified sets of nonscale parameters. One simple way to achieve this is to make ![]() and
and ![]() ; that is, you simply equate the scale parameters of both components and keep the set of nonscale parameters unchanged. However,
forcing the scale parameters to be equal in both components is restrictive, because the mixture cannot model potential differences
in the scales of the two components. A better approach is to tie the scale parameters of the two components by a ratio such
that
; that is, you simply equate the scale parameters of both components and keep the set of nonscale parameters unchanged. However,
forcing the scale parameters to be equal in both components is restrictive, because the mixture cannot model potential differences
in the scales of the two components. A better approach is to tie the scale parameters of the two components by a ratio such
that ![]() and
and ![]() . If the ratio parameter
. If the ratio parameter ![]() is estimated along with the other parameters, then the mixture distribution becomes flexible enough to model the variations
across the scale parameters of individual components.
is estimated along with the other parameters, then the mixture distribution becomes flexible enough to model the variations
across the scale parameters of individual components.
To summarize, the PDF and CDF are of the following form for the two-component mixture that has a scale parameter:
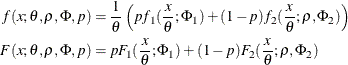
This can be generalized to a mixture of K components by introducing the ![]() ratio parameters
ratio parameters ![]() that relate the scale parameters of each of the K components to the scale parameter
that relate the scale parameters of each of the K components to the scale parameter ![]() of the mixture distribution as follows:
of the mixture distribution as follows:
In order to illustrate this approach, define a mixture of two lognormal distributions by using the following PDF function:
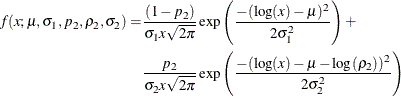
You can verify that ![]() serves as the log of the scale parameter
serves as the log of the scale parameter ![]() (
(![]() ).
).
The following PROC FCMP steps encode this formulation in a distribution named SLOGNMIX2 for use with PROC HPSEVERITY:
/*- Define Mixture of 2 Lognormal Distributions with a Log-Scale Parameter -*/
proc fcmp library=sashelp.svrtdist outlib=work.sevexmpl.models;
function slognmix2_description() $128;
return ("Mixture of two lognormals with a log-scale parameter Mu");
endsub;
function slognmix2_scaletransform() $8;
return ("LOG");
endsub;
function slognmix2_pdf(x, Mu, Sigma1, p2, Rho2, Sigma2);
Mu1 = Mu;
Mu2 = Mu + log(Rho2);
pdf1 = logn_pdf(x, Mu1, Sigma1);
pdf2 = logn_pdf(x, Mu2, Sigma2);
return ((1-p2)*pdf1 + p2*pdf2);
endsub;
function slognmix2_cdf(x, Mu, Sigma1, p2, Rho2, Sigma2);
Mu1 = Mu;
Mu2 = Mu + log(Rho2);
cdf1 = logn_cdf(x, Mu1, Sigma1);
cdf2 = logn_cdf(x, Mu2, Sigma2);
return ((1-p2)*cdf1 + p2*cdf2);
endsub;
subroutine slognmix2_parminit(dim, x[*], nx[*], F[*], Ftype,
Mu, Sigma1, p2, Rho2, Sigma2);
outargs Mu, Sigma1, p2, Rho2, Sigma2;
array m[1] / nosymbols;
p2 = 0.5;
Rho2 = 0.5;
median = svrtutil_percentile(0.5, dim, x, F, Ftype);
Mu = log(2*median/1.5);
call svrtutil_rawmoments(dim, x, nx, 1, m);
lm1 = log(m[1]);
/* Search Rho2 that makes log(sample mean) > Mu */
do while (lm1 <= Mu and Rho2 < 1);
Rho2 = Rho2 + 0.01;
Mu = log(2*median/(1+Rho2));
end;
if (Rho2 >= 1) then
/* If Mu cannot be decreased enough to make it less
than log(sample mean), then revert to Rho2=0.5.
That will set Sigma1 and possibly Sigma2 to missing.
PROC HPSEVERITY replaces missing initial values with 0.001. */
Mu = log(2*median/1.5);
Sigma1 = sqrt(2.0*(log(m[1])-Mu));
Sigma2 = sqrt(2.0*(log(m[1])-Mu-log(Rho2)));
endsub;
subroutine slognmix2_lowerbounds(Mu, Sigma1, p2, Rho2, Sigma2);
outargs Mu, Sigma1, p2, Rho2, Sigma2;
Mu = .; /* Mu has no lower bound */
Sigma1 = 0; /* Sigma1 > 0 */
p2 = 0; /* p2 > 0 */
Rho2 = 0; /* Rho2 > 0 */
Sigma2 = 0; /* Sigma2 > 0 */
endsub;
subroutine slognmix2_upperbounds(Mu, Sigma1, p2, Rho2, Sigma2);
outargs Mu, Sigma1, p2, Rho2, Sigma2;
Mu = .; /* Mu has no upper bound */
Sigma1 = .; /* Sigma1 has no upper bound */
p2 = 1; /* p2 < 1 */
Rho2 = 1; /* Rho2 < 1 */
Sigma2 = .; /* Sigma2 has no upper bound */
endsub;
quit;
As shown in previous examples, an important aspect of defining a distribution for use with PROC HPSEVERITY is the definition
of the PARMINIT subroutine that initializes the parameters. For mixture distributions, in general, the parameter initialization
is a nontrivial task. For a two-component mixture, some simplifying assumptions make the problem easier to handle. For the
initialization of SLOGNMIX2, the initial values of ![]() and
and ![]() are fixed at 0.5, and the following two simplifying assumptions are made:
are fixed at 0.5, and the following two simplifying assumptions are made:
-
The median of the mixture is the average of the medians of the two components:
![\[ F^{-1}(0.5) = (\exp (\mu _1) + \exp (\mu _2))/2 = \exp (\mu ) (1+\rho _2)/2 \]](images/etshpug_hpseverity0711.png)
Solution of this equation yields the value of
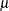 in terms of
in terms of 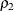 and the sample median.
and the sample median.
-
Each component has the same mean, which implies the following:
![\[ \exp (\mu + \sigma _1^2/2) = \exp (\mu + \log (\rho _2) + \sigma _2^2/2) \]](images/etshpug_hpseverity0712.png)
If
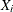 represents the random variable of component distribution i and X represents the random variable of the mixture distribution, then the following equation holds for the raw moment of any order
k:
represents the random variable of component distribution i and X represents the random variable of the mixture distribution, then the following equation holds for the raw moment of any order
k:
![\[ E[X^ k] = \sum _{i=1}^{K} p_ i E[X_ i^ k] \]](images/etshpug_hpseverity0714.png)
This, in conjunction with the assumption on component means, leads to the equations
![\[ \log (m_1) = \mu + \frac{\sigma _1^2}{2} \\ \log (m_1) = \mu + \log (\rho _2) + \frac{\sigma _2^2}{2} \\ \]](images/etshpug_hpseverity0715.png)
where
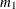 denotes the first raw moment of the sample. Solving these equations leads to the following values of
denotes the first raw moment of the sample. Solving these equations leads to the following values of 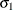 and
and 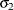 :
:
![\[ \sigma _1^2 = 2 (\log (m_1) - \mu ) \\ \sigma _2^2 = 2 (\log (m_1) - \mu - \log (\rho _2)) \\ \]](images/etshpug_hpseverity0718.png)
Note that
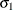 has a valid value only if
has a valid value only if 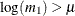 . Among the many possible methods of ensuring this condition, the SLOGNMIX2_PARMINIT subroutine uses the method of doing a
linear search over
. Among the many possible methods of ensuring this condition, the SLOGNMIX2_PARMINIT subroutine uses the method of doing a
linear search over 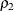 .
.
Even when the preceding assumptions are not true for a given problem, they produce reasonable initial values to help guide the nonlinear optimizer to an acceptable optimum if the mixture of two lognormal distributions is indeed a good fit for your input data. This is illustrated by the results of the following steps that fit the SLOGNMIX2 distribution to simulated data, which have different means for the two components (12.18 and 22.76, respectively), and the median of the sample (15.94) is not equal to the average of the medians of the two components (7.39 and 20.09, respectively):
/*-------- Simulate a lognormal mixture sample ----------*/
data testlognmix(keep=y);
call streaminit(12345);
Mu1 = 2;
Sigma1 = 1;
i = 0;
do j=1 to 2000;
y = exp(Mu1) * rand('LOGNORMAL')**Sigma1;
output;
end;
Mu2 = 3;
Sigma2 = 0.5;
do j=1 to 3000;
y = exp(Mu2) * rand('LOGNORMAL')**Sigma2;
output;
end;
run;
/*-- Fit and compare scale regression models with 2-component --*/
/*-- lognormal mixture and the standard lognormal distribution --*/
options cmplib=(work.sevexmpl);
proc hpseverity data=testlognmix print=all;
loss y;
dist slognmix2 logn;
run;
The comparison of the fit statistics of SLOGNMIX2 and LOGN, as shown in Output 9.8.1, confirms that the two-component mixture is certainly a better fit to these data than the single lognormal distribution.
Output 9.8.1: Comparison of Fitting One versus Two Lognormal Components to Mixture Data
| All Fit Statistics | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Distribution | -2 Log Likelihood |
AIC | AICC | BIC | KS | AD | CvM | |||||||
| slognmix2 | 38343 | * | 38353 | * | 38353 | * | 38386 | * | 0.52221 | * | 0.19843 | * | 0.02728 | * |
| Logn | 39073 | 39077 | 39077 | 39090 | 5.86522 | 66.93414 | 11.72703 | |||||||
| Note: The asterisk (*) marks the best model according to each column's criterion. | ||||||||||||||
The detailed results for the SLOGNMIX2 distribution are shown in Output 9.8.2. According to the "Initial Parameter Values and Bounds" table, the initial value of ![]() is not 0.5, indicating that a linear search was conducted to ensure
is not 0.5, indicating that a linear search was conducted to ensure ![]() .
.
By using the relationship that ![]() , you can see that the final parameter estimates are indeed close to the true parameter values that were used to simulate
the input sample.
, you can see that the final parameter estimates are indeed close to the true parameter values that were used to simulate
the input sample.