The PROC HPQLIM statement invokes the HPQLIM procedure. You can specify the following options.
-
DATA=SAS-data-set
-
specifies the input SAS data set. If this option is not specified, PROC HPQLIM uses the most recently created SAS data set.
-
OUTEST=SAS-data-set
-
writes the parameter estimates to an output data set.
-
COVOUT
-
writes the covariance matrix for the parameter estimates to the OUTEST= data set. This option is valid only if the OUTEST=
option is specified.
-
CORROUT
-
writes the correlation matrix for the parameter estimates to the OUTEST= data set. This option is valid only if the OUTEST=
option is specified.
-
NOPRINT
-
suppresses the normal printed output but does not suppress error listings. If this option is specified, then any other print
option is turned off.
-
PRINTALL
-
turns on all the printing options. The options that are set by PRINTALL are COVB and CORRB.
-
CORRB
-
prints the correlation matrix of the parameter estimates.
-
COVB
-
prints the covariance matrix of the parameter estimates.
-
COVEST=covariance-option
-
specifies the method for calculating the covariance matrix of parameter estimates. You can specify the following covariance-options.
- OP
-
specifies the covariance from the outer product matrix.
- HESSIAN
-
specifies the covariance from the inverse Hessian matrix.
- QML
-
specifies the covariance from the outer product and Hessian matrices (the quasi-maximum likelihood estimates).
The default is COVEST=HESSIAN.
Optimization Control Options
PROC HPQLIM uses the nonlinear optimization (NLO) subsystem to perform nonlinear optimization tasks. You can specify the following
options:
-
ABSCONV=r
ABSTOL=r
-
specifies an absolute function value convergence criterion by which minimization stops when 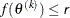 . The default value of r is the negative square root of the largest double-precision value, which serves only as a protection against overflows.
. The default value of r is the negative square root of the largest double-precision value, which serves only as a protection against overflows.
-
ABSFCONV=r
ABSFTOL=r
-
specifies an absolute function difference convergence criterion by which minimization stops when the function value has a
small change in successive iterations:
The default value is 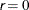 .
.
-
ABSGCONV=r
ABSGTOL=r
-
specifies an absolute gradient convergence criterion. Optimization stops when the maximum absolute gradient element is small:
The default value is r=1E–5.
-
ABSXCONV=r
ABSXTOL=r
-
specifies an absolute parameter convergence criterion. Optimization stops when the Euclidean distance between successive parameter
vectors is small:
The default is 0.
-
FCONV=r
FTOL=r
-
specifies a relative function convergence criterion. Optimization stops when a relative change of the function value in successive
iterations is small:
The default value is 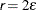 , where
, where  denotes the machine precision constant, which is the smallest double-precision floating-point number such that
denotes the machine precision constant, which is the smallest double-precision floating-point number such that 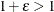 .
.
-
GCONV=r
GTOL=r
-
specifies a relative gradient convergence criterion. For all techniques except CONGRA, optimization stops when the normalized
predicted function reduction is small:
For the CONGRA technique (where a reliable Hessian estimate 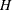 is not available), the following criterion is used:
is not available), the following criterion is used:
The default value is 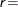 1E–8.
1E–8.
-
MAXFUNC=i
MAXFU=i
-
specifies the maximum number of function calls in the optimization process. The default is 1,000.
The optimization can terminate only after completing a full iteration. Therefore, the number of function calls that are actually
performed can exceed the number of calls that are specified by this option.
-
MAXITER=i
MAXIT=i
-
specifies the maximum number of iterations in the optimization process. The default is 200.
-
MAXTIME=r
-
specifies an upper limit of r seconds of CPU time for the optimization process. The default value is the largest floating-point double representation of
your computer. The time that is specified by this option is checked only once at the end of each iteration. Therefore, the
actual running time can be much longer than r. The actual running time includes the remaining time needed to finish the iteration and the time needed to generate the output
of the results.
-
METHOD=value
-
specifies the iterative minimization method to use. The default is METHOD=NEWRAP. You can specify the following values:
- CONGRA
-
specifies the conjugate-gradient method.
- DBLDOG
-
specifies the double dogleg method.
- NONE
-
specifies that no optimization be performed beyond using the ordinary least squares method to compute the parameter estimates.
- NEWRAP
-
specifies the Newton-Raphson method (the default).
- NRRIDG
-
specifies the Newton-Raphson Ridge method.
- QUANEW
-
specifies the quasi-Newton method.
- TRUREG
-
specifies the trust region method.
-
SINGULAR=r
-
specifies the general singularity criterion that is applied by the HPQLIM procedure in sweeps and inversions. The default
for the optimization is 1E–8.
-
PLOTS<(global-plot-options)> = plot-request | (plot-requests)
-
controls the display of plots. By default, the plots are displayed in panels unless the UNPACK global-plot-option is specified. When you specify only one plot-request, you can omit the parentheses around it.
You can specify the following global-plot-options:
-
ONLY
-
displays only the requested plot.
-
UNPACKPANEL
UNPACK
-
specifies that all paneled plots be unpacked, meaning that each plot in a panel is displayed separately.
You can specify the following plot-requests:
-
ALL
-
specifies all types of available plots.
-
AUTOCORR<(LAGS=n)>
-
displays the autocorrelation function plots for the parameters. The optional LAGS= suboption specifies the number (up to lag
n) of autocorrelations to be plotted in the autocorrelation function plot. If this suboption is not specified, autocorrelations
are plotted up to lag 50. This plot-request is available only for Bayesian analysis.
-
BAYESDIAG
-
is equivalent to specifying the TRACE, AUTOCORR, and DENSITY plot-requests.
-
DENSITY<(FRINGE)>
-
displays the kernel density plots for the parameters. If you specify the FRINGE suboption, a fringe plot is created on the
X axis of the kernel density plot. This plot-request is available only for Bayesian analysis.
-
NONE
-
suppresses all diagnostic plots.
-
TRACE<(SMOOTH)>
-
displays the trace plots for the parameters. The SMOOTH suboption displays a fitted penalized B-spline curve for each plot.
This plot-request is available only for Bayesian analysis.
Copyright © SAS Institute Inc. All Rights Reserved.
