The LIFEREG Procedure
- Overview
-
Getting Started

-
Syntax

-
Details
 Missing ValuesModel SpecificationComputational MethodSupported DistributionsPredicted ValuesConfidence IntervalsFit StatisticsProbability PlottingINEST= Data SetOUTEST= Data SetXDATA= Data SetComputational ResourcesBayesian AnalysisDisplayed Output for Classical AnalysisDisplayed Output for Bayesian AnalysisODS Table NamesODS Graphics
Missing ValuesModel SpecificationComputational MethodSupported DistributionsPredicted ValuesConfidence IntervalsFit StatisticsProbability PlottingINEST= Data SetOUTEST= Data SetXDATA= Data SetComputational ResourcesBayesian AnalysisDisplayed Output for Classical AnalysisDisplayed Output for Bayesian AnalysisODS Table NamesODS Graphics -
Examples
 Motorette FailureComputing Predicted Values for a Tobit ModelOvercoming Convergence Problems by Specifying Initial ValuesAnalysis of Arbitrarily Censored Data with Interaction EffectsProbability Plotting—Right CensoringProbability Plotting—Arbitrary CensoringBayesian Analysis of Clinical Trial DataModel Postfitting Analysis
Motorette FailureComputing Predicted Values for a Tobit ModelOvercoming Convergence Problems by Specifying Initial ValuesAnalysis of Arbitrarily Censored Data with Interaction EffectsProbability Plotting—Right CensoringProbability Plotting—Arbitrary CensoringBayesian Analysis of Clinical Trial DataModel Postfitting Analysis - References
Bayesian Analysis
Gibbs Sampling
This section provides details about Bayesian analysis by Gibbs sampling in the location-scale models for survival data available in PROC LIFEREG. See the section Gibbs Sampler for a general discussion of Gibbs sampling. PROC LIFEREG fits parametric location-scale survival models. That is, the probability density of the response Y can expressed in the general form
|
|
where ![]() for lifetimes T. The function g determines the specific distribution. The location parameter
for lifetimes T. The function g determines the specific distribution. The location parameter ![]() is modeled through regression parameters as
is modeled through regression parameters as ![]() . The LIFEREG procedure can provide Bayesian estimates of the regression parameters and
. The LIFEREG procedure can provide Bayesian estimates of the regression parameters and ![]() . The OUTPUT and PROBPLOT statements, if specified, are ignored. The PLOTS=PROBPLOT option in the PROC LIFEREG statement and
the CORRB and COVB options in the MODEL statement are also ignored.
. The OUTPUT and PROBPLOT statements, if specified, are ignored. The PLOTS=PROBPLOT option in the PROC LIFEREG statement and
the CORRB and COVB options in the MODEL statement are also ignored.
For the Weibull distribution, you can specify that Gibbs sampling be performed on the Weibull shape parameter ![]() instead of the scale parameter
instead of the scale parameter ![]() by specifying a prior distribution for the shape parameter with the WEIBULLSHAPEPRIOR= option. In addition, if there are
no covariates in the model, you can specify Gibbs sampling on the Weibull scale parameter
by specifying a prior distribution for the shape parameter with the WEIBULLSHAPEPRIOR= option. In addition, if there are
no covariates in the model, you can specify Gibbs sampling on the Weibull scale parameter ![]() , where
, where ![]() is the intercept term, with the WEIBULLSCALEPRIOR= option.
is the intercept term, with the WEIBULLSCALEPRIOR= option.
In the case of the exponential distribution with no covariates, you can specify Gibbs sampling on the exponential scale parameter
![]() , where
, where ![]() is the intercept term, with the EXPSCALEPRIOR= option.
is the intercept term, with the EXPSCALEPRIOR= option.
Let ![]() be the parameter vector. For location-scale models, the
be the parameter vector. For location-scale models, the ![]() ’s are the regression coefficients
’s are the regression coefficients ![]() ’s and the scale parameter
’s and the scale parameter ![]() . In the case of the three-parameter gamma distribution, there is an additional gamma shape parameter
. In the case of the three-parameter gamma distribution, there is an additional gamma shape parameter ![]() . Let
. Let ![]() be the likelihood function, where D is the observed data. Let
be the likelihood function, where D is the observed data. Let ![]() be the prior distribution. The full conditional distribution of
be the prior distribution. The full conditional distribution of ![]() is proportional to the joint distribution; that is,
is proportional to the joint distribution; that is,
|
|
For instance, the one-dimensional conditional distribution of ![]() given
given ![]() , is computed as
, is computed as
|
|
Suppose you have a set of arbitrary starting values ![]() . Using the ARMS (adaptive rejection Metropolis sampling) algorithm of Gilks and Wild (1992) and Gilks, Best, and Tan (1995), you can do the following:
. Using the ARMS (adaptive rejection Metropolis sampling) algorithm of Gilks and Wild (1992) and Gilks, Best, and Tan (1995), you can do the following:
-
draw
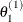 from
from ![$[\theta _1|\theta _2^{(0)},\ldots ,\theta _ k^{(0)}]$](images/statug_lifereg0321.png)
-
draw
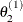 from
from ![$[\theta _2|\theta _1^{(1)}, \theta _3^{(0)},\ldots ,\theta _ k^{(0)}]$](images/statug_lifereg0323.png)
-

-
draw
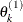 from
from ![$[\theta _ k|\theta _1^{(1)},\ldots ,\theta _{k-1}^{(1)}]$](images/statug_lifereg0326.png)
This completes one iteration of the Gibbs sampler. After one iteration, you have ![]() . After n iterations, you have
. After n iterations, you have ![]() . PROC LIFEREG implements the ARMS algorithm based on a program provided by Gilks (2003) to draw a sample from a full conditional distribution. See the section Assessing Markov Chain Convergence for information about assessing the convergence of the chain of posterior samples.
. PROC LIFEREG implements the ARMS algorithm based on a program provided by Gilks (2003) to draw a sample from a full conditional distribution. See the section Assessing Markov Chain Convergence for information about assessing the convergence of the chain of posterior samples.
You can output these posterior samples into a SAS data set. The following option in the BAYES statement outputs the posterior
samples into the SAS data set Post: OUTPOST=Post. The data set also includes the variables LogPost and LogLike, which represent the log of the posterior distribution and the log of the likelihood, respectively.
Priors for Model Parameters
The model parameters are the regression coefficients and the dispersion parameter (or the precision or scale), if the model has one. The priors for the dispersion parameter and the priors for the regression coefficients are assumed to be independent, while you can have a joint multivariate normal prior for the regression coefficients.
Scale and Shape Parameters
Gamma Prior
The gamma distribution ![]() has a PDF
has a PDF
|
|
where a is the shape parameter and b is the inverse-scale parameter. The mean is ![]() and the variance is
and the variance is ![]() .
.
Improper Prior
The joint prior density is given by
|
|
Regression Coefficients
Let ![]() be the regression coefficients.
be the regression coefficients.
Normal Prior
Assume ![]() has a multivariate normal prior with mean vector
has a multivariate normal prior with mean vector ![]() and covariance matrix
and covariance matrix ![]() . The joint prior density is given by
. The joint prior density is given by
|
|
Uniform Prior
The joint prior density is given by
|
|
Posterior Distribution
Denote the observed data by D.
The posterior distribution is
|
|
where ![]() is the likelihood function with regression coefficients and any additional parameters, such as scale or shape,
is the likelihood function with regression coefficients and any additional parameters, such as scale or shape, ![]() as parameters; and
as parameters; and ![]() is the joint prior distribution of the parameters.
is the joint prior distribution of the parameters.
Deviance Information Criterion
Let ![]() be the model parameters at iteration i of the Gibbs sampler, and let LL(
be the model parameters at iteration i of the Gibbs sampler, and let LL(![]() ) be the corresponding model log likelihood. PROC LIFEREG computes the following fit statistics defined by Spiegelhalter et al.
(2002):
) be the corresponding model log likelihood. PROC LIFEREG computes the following fit statistics defined by Spiegelhalter et al.
(2002):
-
effective number of parameters:
![\[ p_ D=\overline{\mr {LL}(\btheta )} - \mr {LL}(\bar{\btheta }) \]](images/statug_lifereg0342.png)
-
deviance information criterion (DIC):
![\[ \mr {DIC}= \overline{\mr {LL}(\btheta )} + p_ D \]](images/statug_lifereg0343.png)
where
|
|
|
|
and n is the number of Gibbs samples.
Starting Values of the Markov Chains
When the BAYES statement is specified, PROC LIFEREG generates one Markov chain containing the approximate posterior samples of the model parameters. Additional chains are produced when the Gelman-Rubin diagnostics are requested. Starting values (or initial values) can be specified in the INITIAL= data set in the BAYES statement. If INITIAL= option is not specified, PROC LIFEREG picks its own initial values for the chains.
Denote ![]() as the integral value of x. Denote
as the integral value of x. Denote ![]() as the estimated standard error of the estimator X.
as the estimated standard error of the estimator X.
Regression Coefficients and Gamma Shape Parameter
For the first chain that the summary statistics and regression diagnostics are based on, the default initial values are estimates of the mode of the posterior distribution. If the INITIALMLE option is specified, the initial values are the maximum likelihood estimates; that is,
|
|
Initial values for the rth chain (![]() ) are given by
) are given by
|
|
with the plus sign for odd r and minus sign for even r.
Scale, Exponential Scale, Weibull Scale, or Weibull Shape Parameter 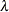
Let ![]() be the parameter sampled.
be the parameter sampled.
For the first chain that the summary statistics and diagnostics are based on, the initial values are estimates of the mode of the posterior distribution; or the maximum likelihood estimates if the INITIALMLE option is specified; that is,
|
|
The initial values of the rth chain (![]() ) are given by
) are given by
|
|
with the plus sign for odd r and minus sign for even r.
OUTPOST= Output Data Set
The OUTPOST= data set contains the generated posterior samples. There are 2+n variables, where n is the number of model parameters. The variable Iteration represents the iteration number and the variable LogPost contains the log posterior likelihood values. The other n variables represent the draws of the Markov chain for the model parameters.