The GENMOD Procedure
-
Overview

-
Getting Started

-
Syntax
 PROC GENMOD StatementASSESS StatementBAYES StatementBY StatementCLASS StatementCODE StatementCONTRAST StatementDEVIANCE StatementEFFECTPLOT StatementESTIMATE StatementEXACT StatementEXACTOPTIONS StatementFREQ StatementFWDLINK StatementINVLINK StatementLSMEANS StatementLSMESTIMATE StatementMODEL StatementOUTPUT StatementProgramming StatementsREPEATED StatementSLICE StatementSTORE StatementSTRATA StatementVARIANCE StatementWEIGHT StatementZEROMODEL Statement
PROC GENMOD StatementASSESS StatementBAYES StatementBY StatementCLASS StatementCODE StatementCONTRAST StatementDEVIANCE StatementEFFECTPLOT StatementESTIMATE StatementEXACT StatementEXACTOPTIONS StatementFREQ StatementFWDLINK StatementINVLINK StatementLSMEANS StatementLSMESTIMATE StatementMODEL StatementOUTPUT StatementProgramming StatementsREPEATED StatementSLICE StatementSTORE StatementSTRATA StatementVARIANCE StatementWEIGHT StatementZEROMODEL Statement -
Details
 Generalized Linear Models TheorySpecification of EffectsParameterization Used in PROC GENMODType 1 AnalysisType 3 AnalysisConfidence Intervals for ParametersF StatisticsLagrange Multiplier StatisticsPredicted Values of the MeanResidualsMultinomial ModelsZero-Inflated ModelsGeneralized Estimating EquationsAssessment of Models Based on Aggregates of ResidualsCase Deletion Diagnostic StatisticsBayesian AnalysisExact Logistic and Exact Poisson RegressionMissing ValuesDisplayed Output for Classical AnalysisDisplayed Output for Bayesian AnalysisDisplayed Output for Exact AnalysisODS Table NamesODS Graphics
Generalized Linear Models TheorySpecification of EffectsParameterization Used in PROC GENMODType 1 AnalysisType 3 AnalysisConfidence Intervals for ParametersF StatisticsLagrange Multiplier StatisticsPredicted Values of the MeanResidualsMultinomial ModelsZero-Inflated ModelsGeneralized Estimating EquationsAssessment of Models Based on Aggregates of ResidualsCase Deletion Diagnostic StatisticsBayesian AnalysisExact Logistic and Exact Poisson RegressionMissing ValuesDisplayed Output for Classical AnalysisDisplayed Output for Bayesian AnalysisDisplayed Output for Exact AnalysisODS Table NamesODS Graphics -
Examples
 Logistic RegressionNormal Regression, Log Link Gamma Distribution Applied to Life DataOrdinal Model for Multinomial DataGEE for Binary Data with Logit Link FunctionLog Odds Ratios and the ALR AlgorithmLog-Linear Model for Count DataModel Assessment of Multiple Regression Using Aggregates of ResidualsAssessment of a Marginal Model for Dependent DataBayesian Analysis of a Poisson Regression ModelExact Poisson Regression
Logistic RegressionNormal Regression, Log Link Gamma Distribution Applied to Life DataOrdinal Model for Multinomial DataGEE for Binary Data with Logit Link FunctionLog Odds Ratios and the ALR AlgorithmLog-Linear Model for Count DataModel Assessment of Multiple Regression Using Aggregates of ResidualsAssessment of a Marginal Model for Dependent DataBayesian Analysis of a Poisson Regression ModelExact Poisson Regression - References
Bayesian Analysis
In generalized linear models, the response has a probability distribution from a family of distributions of the exponential form. That is, the probability density of the response Y for continuous response variables, or the probability function for discrete responses, can be expressed as
|
|
for some functions a, b, and c that determine the specific distribution. The canonical parameters ![]() depend only on the means of the response
depend only on the means of the response ![]() , which are related to the regression parameters
, which are related to the regression parameters ![]() through the link function
through the link function ![]() . The additional parameter
. The additional parameter ![]() is the dispersion parameter. The GENMOD procedure estimates the regression parameters and the scale parameter
is the dispersion parameter. The GENMOD procedure estimates the regression parameters and the scale parameter ![]() by maximum likelihood. However, the GENMOD procedure can also provide Bayesian estimates of the regression parameters and
either the scale
by maximum likelihood. However, the GENMOD procedure can also provide Bayesian estimates of the regression parameters and
either the scale ![]() , the dispersion
, the dispersion ![]() , or the precision
, or the precision ![]() by sampling from the posterior distribution. Except where noted, the following discussion applies to either
by sampling from the posterior distribution. Except where noted, the following discussion applies to either ![]() ,
, ![]() , or
, or ![]() , although
, although ![]() is used to illustrate the formulas. Note that the Poisson and binomial distributions do not have a dispersion parameter,
and the dispersion is considered to be fixed at
is used to illustrate the formulas. Note that the Poisson and binomial distributions do not have a dispersion parameter,
and the dispersion is considered to be fixed at ![]() . The ASSESS, CONTRAST, ESTIMATE, OUTPUT, and REPEATED statements, if specified, are ignored. Also ignored are the PLOTS=
option in the PROC GENMOD statement and the following options in the MODEL statement: ALPHA=, CORRB, COVB, TYPE1, TYPE3, SCALE=DEVIANCE
(DSCALE), SCALE=PEARSON (PSCALE), OBSTATS, RESIDUALS, XVARS, PREDICTED, DIAGNOSTICS, and SCALE= for Poisson and binomial distributions.
The multinomial and zero-inflated Poisson distributions are not available for Bayesian analysis.
. The ASSESS, CONTRAST, ESTIMATE, OUTPUT, and REPEATED statements, if specified, are ignored. Also ignored are the PLOTS=
option in the PROC GENMOD statement and the following options in the MODEL statement: ALPHA=, CORRB, COVB, TYPE1, TYPE3, SCALE=DEVIANCE
(DSCALE), SCALE=PEARSON (PSCALE), OBSTATS, RESIDUALS, XVARS, PREDICTED, DIAGNOSTICS, and SCALE= for Poisson and binomial distributions.
The multinomial and zero-inflated Poisson distributions are not available for Bayesian analysis.
See the section Assessing Markov Chain Convergence for information about assessing the convergence of the chain of posterior samples.
Several algorithms, specified with the SAMPLING= option in the BAYES statement, are available in GENMOD for drawing samples from the posterior distribution.
ARMS Algorithm for Gibbs Sampling
This section provides details for Bayesian analysis by Gibbs sampling in generalized linear models. See the section Gibbs Sampler for a general discussion of Gibbs sampling. See Gilks, Richardson, and Spiegelhalter (1996) for a discussion of applications of Gibbs sampling to a number of different models, including generalized linear models.
Let ![]() be the parameter vector. For generalized linear models, the
be the parameter vector. For generalized linear models, the ![]() s are the regression coefficients
s are the regression coefficients ![]() s and the dispersion parameter
s and the dispersion parameter ![]() . Let
. Let ![]() be the likelihood function, where D is the observed data. Let
be the likelihood function, where D is the observed data. Let ![]() be the prior distribution. The full conditional distribution of
be the prior distribution. The full conditional distribution of ![]() is proportional to the joint distribution; that is,
is proportional to the joint distribution; that is,
|
|
For instance, the one-dimensional conditional distribution of ![]() given
given ![]() , is computed as
, is computed as
|
|
Suppose you have a set of arbitrary starting values ![]() . Using the ARMS (adaptive rejection Metropolis sampling) algorithm (Gilks and Wild, 1992; Gilks, Best, and Tan, 1995), you can do the following:
. Using the ARMS (adaptive rejection Metropolis sampling) algorithm (Gilks and Wild, 1992; Gilks, Best, and Tan, 1995), you can do the following:
-
draw
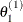 from
from ![$[\theta _1|\theta _2^{(0)},\ldots ,\theta _ k^{(0)}]$](images/statug_genmod0631.png)
-
draw
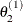 from
from ![$[\theta _2|\theta _1^{(1)}, \theta _3^{(0)},\ldots ,\theta _ k^{(0)}]$](images/statug_genmod0633.png)
-

-
draw
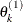 from
from ![$[\theta _ k|\theta _1^{(1)},\ldots ,\theta _{k-1}^{(1)}]$](images/statug_genmod0636.png)
This completes one iteration of the Gibbs sampler. After one iteration, you have ![]() . After n iterations, you have
. After n iterations, you have ![]() . PROC GENMOD implements the ARMS algorithm provided by Gilks (2003) to draw a sample from a full conditional distribution. See the section Adaptive Rejection Sampling Algorithm for more information about the ARMS algorithm. The ARMS algorithm is the default method used to sample from the posterior
distribution, except in the case of a normal distribution with a conjugate prior, in which case a closed form is available
for the posterior distribution. See any of the introductory references in Chapter 7: Introduction to Bayesian Analysis Procedures, for a discussion of conjugate prior distributions for a linear model with the normal distribution.
. PROC GENMOD implements the ARMS algorithm provided by Gilks (2003) to draw a sample from a full conditional distribution. See the section Adaptive Rejection Sampling Algorithm for more information about the ARMS algorithm. The ARMS algorithm is the default method used to sample from the posterior
distribution, except in the case of a normal distribution with a conjugate prior, in which case a closed form is available
for the posterior distribution. See any of the introductory references in Chapter 7: Introduction to Bayesian Analysis Procedures, for a discussion of conjugate prior distributions for a linear model with the normal distribution.
Gamerman Algorithm
The Gamerman algorithm, unlike a Gibbs sampling algorithm, samples parameters from their multivariate posterior conditional distribution. The algorithm uses the structure of generalized linear models to efficiently sample from the posterior distribution of the model parameters. For a detailed description and explanation of the algorithm, see Gamerman (1997) and the section Gamerman Algorithm.
Independence Metropolis Algorithm
The independence Metropolis algorithm is another sampling algorithm that draws multivariate samples from the posterior distribution. See the section Independence Sampler for more details.
Posterior Samples Output Data Set
You can output posterior samples into a SAS data set through ODS. The following SAS statement outputs the posterior samples
into the SAS data set Post:
ODS OUTPUT POSTERIORSAMPLE=Post
You can alternatively create the SAS data set Post with the OUTPOST=Post option in the BAYES statement.
The data set also includes the variables LogPost and LogLike, which represent the log of the posterior likelihood and the log of the likelihood, respectively.
Priors for Model Parameters
The model parameters are the regression coefficients and the dispersion parameter (or the precision or scale), if the model has one. The priors for the dispersion parameter and the priors for the regression coefficients are assumed to be independent, while you can have a joint multivariate normal prior for the regression coefficients.
Dispersion, Precision, or Scale Parameter
Gamma Prior
The gamma distribution ![]() has a probability density function
has a probability density function
|
|
where a is the shape parameter and b is the inverse-scale parameter. The mean is ![]() and the variance is
and the variance is ![]() .
.
Improper Prior
The joint prior density is given by
|
|
Inverse Gamma Prior
The inverse gamma distribution ![]() has a probability density function
has a probability density function
|
|
where a is the shape parameter and b is the scale parameter. The mean is ![]() if
if ![]() , and the variance is
, and the variance is ![]() if
if ![]() .
.
Regression Coefficients
Let ![]() be the regression coefficients.
be the regression coefficients.
Jeffreys’ Prior
The joint prior density is given by
|
|
where ![]() is the Fisher information matrix for the model. If the underlying model has a scale parameter (for example, a normal linear
regression model), then the Fisher information matrix is computed with the scale parameter set to a fixed value of one.
is the Fisher information matrix for the model. If the underlying model has a scale parameter (for example, a normal linear
regression model), then the Fisher information matrix is computed with the scale parameter set to a fixed value of one.
If you specify the CONDITIONAL option, then Jeffreys’ prior, conditional on the current Markov chain value of the generalized
linear model precision parameter ![]() , is given by
, is given by
|
|
where ![]() is the model precision parameter.
is the model precision parameter.
See Ibrahim and Laud (1991) for a full discussion, with examples, of Jeffreys’ prior for generalized linear models.
Normal Prior
Assume ![]() has a multivariate normal prior with mean vector
has a multivariate normal prior with mean vector ![]() and covariance matrix
and covariance matrix ![]() . The joint prior density is given by
. The joint prior density is given by
|
|
If you specify the CONDITIONAL option, then, conditional on the current Markov chain value of the generalized linear model
precision parameter ![]() , the joint prior density is given by
, the joint prior density is given by
|
|
Uniform Prior
The joint prior density is given by
|
|
Deviance Information Criterion
Let ![]() be the model parameters at iteration i of the Gibbs sampler and let LL(
be the model parameters at iteration i of the Gibbs sampler and let LL(![]() ) be the corresponding model log likelihood. PROC GENMOD computes the following fit statistics defined by Spiegelhalter et al.
(2002):
) be the corresponding model log likelihood. PROC GENMOD computes the following fit statistics defined by Spiegelhalter et al.
(2002):
-
Effective number of parameters:
![\[ p_ D=\overline{\mr {LL}(\theta )} - \mr {LL}(\bar{\theta }) \]](images/statug_genmod0656.png)
-
Deviance information criterion (DIC):
![\[ \mr {DIC}= \overline{\mr {LL}(\theta )} + p_ D \]](images/statug_genmod0657.png)
where
|
|
PROC GENMOD uses the full log likelihoods defined in the section Log-Likelihood Functions, with all terms included, for computing the DIC.
Posterior Distribution
Denote the observed data by D.
The posterior distribution is
|
|
where ![]() is the likelihood function with regression coefficients
is the likelihood function with regression coefficients ![]() as parameters.
as parameters.
Starting Values of the Markov Chains
When the BAYES statement is specified, PROC GENMOD generates one Markov chain containing the approximate posterior samples of the model parameters. Additional chains are produced when the Gelman-Rubin diagnostics are requested. Starting values (or initial values) can be specified in the INITIAL= data set in the BAYES statement. If INITIAL= option is not specified, PROC GENMOD picks its own initial values for the chains.
Denote ![]() as the integral value of x. Denote
as the integral value of x. Denote ![]() as the estimated standard error of the estimator X.
as the estimated standard error of the estimator X.
Regression Coefficients
For the first chain that the summary statistics and regression diagnostics are based on, the default initial values are estimates of the mode of the posterior distribution. If the INITIALMLE option is specified, the initial values are the maximum likelihood estimates; that is,
|
|
Initial values for the rth chain (![]() ) are given by
) are given by
|
|
with the plus sign for odd r and minus sign for even r.
Dispersion, Scale, or Precision Parameter 
Let ![]() be the generalized linear model parameter you choose to sample, either the dispersion, scale, or precision parameter. Note
that the Poisson and binomial distributions do not have this additional parameter.
be the generalized linear model parameter you choose to sample, either the dispersion, scale, or precision parameter. Note
that the Poisson and binomial distributions do not have this additional parameter.
For the first chain that the summary statistics and regression diagnostics are based on, the default initial values are estimates of the mode of the posterior distribution. If the INITIALMLE option is specified, the initial values are the maximum likelihood estimates; that is,
|
|
The initial values of the rth chain (![]() ) are given by
) are given by
|
|
with the plus sign for odd r and minus sign for even r.
OUTPOST= Output Data Set
The OUTPOST= data set contains the generated posterior samples. There are 3+n variables, where n is the number of model parameters. The variable Iteration represents the iteration number, the variable LogLike contains the log of the likelihood, and the variable LogPost contains the log of the posterior. The other n variables represent the draws of the Markov chain for the model parameters.