The GLIMMIX Procedure
-
Overview

-
Getting Started

-
Syntax
 PROC GLIMMIX Statement BY Statement CLASS Statement CONTRAST Statement COVTEST Statement EFFECT Statement ESTIMATE Statement FREQ Statement ID Statement LSMEANS Statement LSMESTIMATE Statement MODEL Statement NLOPTIONS Statement OUTPUT Statement PARMS Statement RANDOM Statement SLICE Statement STORE Statement WEIGHT Statement Programming Statements User-Defined Link or Variance Function
PROC GLIMMIX Statement BY Statement CLASS Statement CONTRAST Statement COVTEST Statement EFFECT Statement ESTIMATE Statement FREQ Statement ID Statement LSMEANS Statement LSMESTIMATE Statement MODEL Statement NLOPTIONS Statement OUTPUT Statement PARMS Statement RANDOM Statement SLICE Statement STORE Statement WEIGHT Statement Programming Statements User-Defined Link or Variance Function -
Details
 Generalized Linear Models Theory Generalized Linear Mixed Models Theory GLM Mode or GLMM Mode Statistical Inference for Covariance Parameters Satterthwaite Degrees of Freedom Approximation Empirical Covariance (Sandwich) Estimators Exploring and Comparing Covariance Matrices Processing by Subjects Radial Smoothing Based on Mixed Models Odds and Odds Ratio Estimation Parameterization of Generalized Linear Mixed Models Response-Level Ordering and Referencing Comparing the GLIMMIX and MIXED Procedures Singly or Doubly Iterative Fitting Default Estimation Techniques Default Output Notes on Output Statistics ODS Table Names ODS Graphics
Generalized Linear Models Theory Generalized Linear Mixed Models Theory GLM Mode or GLMM Mode Statistical Inference for Covariance Parameters Satterthwaite Degrees of Freedom Approximation Empirical Covariance (Sandwich) Estimators Exploring and Comparing Covariance Matrices Processing by Subjects Radial Smoothing Based on Mixed Models Odds and Odds Ratio Estimation Parameterization of Generalized Linear Mixed Models Response-Level Ordering and Referencing Comparing the GLIMMIX and MIXED Procedures Singly or Doubly Iterative Fitting Default Estimation Techniques Default Output Notes on Output Statistics ODS Table Names ODS Graphics -
Examples
 Binomial Counts in Randomized Blocks Mating Experiment with Crossed Random Effects Smoothing Disease Rates; Standardized Mortality Ratios Quasi-likelihood Estimation for Proportions with Unknown Distribution Joint Modeling of Binary and Count Data Radial Smoothing of Repeated Measures Data Isotonic Contrasts for Ordered Alternatives Adjusted Covariance Matrices of Fixed Effects Testing Equality of Covariance and Correlation Matrices Multiple Trends Correspond to Multiple Extrema in Profile Likelihoods Maximum Likelihood in Proportional Odds Model with Random Effects Fitting a Marginal (GEE-Type) Model Response Surface Comparisons with Multiplicity Adjustments Generalized Poisson Mixed Model for Overdispersed Count Data Comparing Multiple B-Splines Diallel Experiment with Multimember Random Effects Linear Inference Based on Summary Data
Binomial Counts in Randomized Blocks Mating Experiment with Crossed Random Effects Smoothing Disease Rates; Standardized Mortality Ratios Quasi-likelihood Estimation for Proportions with Unknown Distribution Joint Modeling of Binary and Count Data Radial Smoothing of Repeated Measures Data Isotonic Contrasts for Ordered Alternatives Adjusted Covariance Matrices of Fixed Effects Testing Equality of Covariance and Correlation Matrices Multiple Trends Correspond to Multiple Extrema in Profile Likelihoods Maximum Likelihood in Proportional Odds Model with Random Effects Fitting a Marginal (GEE-Type) Model Response Surface Comparisons with Multiplicity Adjustments Generalized Poisson Mixed Model for Overdispersed Count Data Comparing Multiple B-Splines Diallel Experiment with Multimember Random Effects Linear Inference Based on Summary Data - References
Example 40.9 Testing Equality of Covariance and Correlation Matrices
Fisher’s iris data are widely used in multivariate statistics. They comprise measurements in millimeters of four flower attributes, the length and width of sepals and petals for 50 specimens from each of three species, Iris setosa, I. versicolor, and I. virginica (Fisher 1936).
When modeling multiple attributes from the same specimen, correlations among measurements from the same flower must be taken into account. Unstructured covariance matrices are common in this multivariate setting. Species comparisons can focus on comparisons of mean response, but comparisons of the variation and covariation are also of interest. In this example, the equivalence of covariance and correlation matrices among the species are examined.
The iris data set is available in the Sashelp library. The following step displays the first 10 observations of the iris data in multivariate format—that is, each observation contains multiple response variables. The DATA step that follows creates a data set in univariate form, where each observation corresponds to a single response variable. This is the form needed by the GLIMMIX procedure.
proc print data=Sashelp.iris(obs=10); run;
| Obs | Species | SepalLength | SepalWidth | PetalLength | PetalWidth |
|---|---|---|---|---|---|
| 1 | Setosa | 50 | 33 | 14 | 2 |
| 2 | Setosa | 46 | 34 | 14 | 3 |
| 3 | Setosa | 46 | 36 | 10 | 2 |
| 4 | Setosa | 51 | 33 | 17 | 5 |
| 5 | Setosa | 55 | 35 | 13 | 2 |
| 6 | Setosa | 48 | 31 | 16 | 2 |
| 7 | Setosa | 52 | 34 | 14 | 2 |
| 8 | Setosa | 49 | 36 | 14 | 1 |
| 9 | Setosa | 44 | 32 | 13 | 2 |
| 10 | Setosa | 50 | 35 | 16 | 6 |
data iris_univ;
set sashelp.iris;
retain id 0;
array y (4) SepalLength SepalWidth PetalLength PetalWidth;
id+1;
do var=1 to 4;
response = y{var};
output;
end;
drop SepalLength SepalWidth PetalLength PetalWidth:;
run;
The following GLIMMIX statements fit a model with separate unstructured covariance matrices for each species:
ods select FitStatistics CovParms CovTests; proc glimmix data=iris_univ; class species var id; model response = species*var; random _residual_ / type=un group=species subject=id; covtest homogeneity; run;
The mean function is modeled as a cell-means model that allows for different means for each species and outcome variable. The covariances are modeled directly (R-side) rather than through random effects. The ID variable identifies the individual plant, so that responses from different plants are independent. The GROUP=SPECIES option varies the parameters of the unstructured covariance matrix by species. Hence, this model has 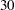 covariance parameters:
covariance parameters: 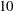 unique parameters for a
unique parameters for a 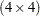 covariance matrix for each of three species.
covariance matrix for each of three species.
The COVTEST statement requests a test of homogeneity—that is, it tests whether varying the covariance parameters by the group effect provides a significantly better fit compared to a model in which different groups share the same parameter.
| Fit Statistics | |
|---|---|
| -2 Res Log Likelihood | 2812.89 |
| AIC (smaller is better) | 2872.89 |
| AICC (smaller is better) | 2876.23 |
| BIC (smaller is better) | 2963.21 |
| CAIC (smaller is better) | 2993.21 |
| HQIC (smaller is better) | 2909.58 |
| Generalized Chi-Square | 588.00 |
| Gener. Chi-Square / DF | 1.00 |
The "Fit Statistics" table shows the 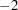 restricted (residual) log likelihood in the full model and other fit statistics (Output 40.9.2). The "-2 Res Log Likelihood" sets the benchmark against which a model with homogeneity constraint is compared. Output 40.9.3 displays the 30 covariance parameters in this model.
restricted (residual) log likelihood in the full model and other fit statistics (Output 40.9.2). The "-2 Res Log Likelihood" sets the benchmark against which a model with homogeneity constraint is compared. Output 40.9.3 displays the 30 covariance parameters in this model.
There appear to be substantial differences among the covariance parameters from different groups. For example, the residual variability of the petal length of the three species is 12.4249, 26.6433, and 40.4343, respectively. The homogeneity hypothesis restricts these variances to be equal and similarly for the other covariance parameters. The results from the COVTEST statement are shown in Output 40.9.4.
| Covariance Parameter Estimates | ||||
|---|---|---|---|---|
| Cov Parm | Subject | Group | Estimate | Standard Error |
| UN(1,1) | id | Species Setosa | 12.4249 | 2.5102 |
| UN(2,1) | id | Species Setosa | 9.9216 | 2.3775 |
| UN(2,2) | id | Species Setosa | 14.3690 | 2.9030 |
| UN(3,1) | id | Species Setosa | 1.6355 | 0.9052 |
| UN(3,2) | id | Species Setosa | 1.1698 | 0.9552 |
| UN(3,3) | id | Species Setosa | 3.0159 | 0.6093 |
| UN(4,1) | id | Species Setosa | 1.0331 | 0.5508 |
| UN(4,2) | id | Species Setosa | 0.9298 | 0.5859 |
| UN(4,3) | id | Species Setosa | 0.6069 | 0.2755 |
| UN(4,4) | id | Species Setosa | 1.1106 | 0.2244 |
| UN(1,1) | id | Species Versicolor | 26.6433 | 5.3828 |
| UN(2,1) | id | Species Versicolor | 8.5184 | 2.6144 |
| UN(2,2) | id | Species Versicolor | 9.8469 | 1.9894 |
| UN(3,1) | id | Species Versicolor | 18.2898 | 4.3398 |
| UN(3,2) | id | Species Versicolor | 8.2653 | 2.4149 |
| UN(3,3) | id | Species Versicolor | 22.0816 | 4.4612 |
| UN(4,1) | id | Species Versicolor | 5.5780 | 1.6617 |
| UN(4,2) | id | Species Versicolor | 4.1204 | 1.0641 |
| UN(4,3) | id | Species Versicolor | 7.3102 | 1.6891 |
| UN(4,4) | id | Species Versicolor | 3.9106 | 0.7901 |
| UN(1,1) | id | Species Virginica | 40.4343 | 8.1690 |
| UN(2,1) | id | Species Virginica | 9.3763 | 3.2213 |
| UN(2,2) | id | Species Virginica | 10.4004 | 2.1012 |
| UN(3,1) | id | Species Virginica | 30.3290 | 6.6262 |
| UN(3,2) | id | Species Virginica | 7.1380 | 2.7395 |
| UN(3,3) | id | Species Virginica | 30.4588 | 6.1536 |
| UN(4,1) | id | Species Virginica | 4.9094 | 2.5916 |
| UN(4,2) | id | Species Virginica | 4.7629 | 1.4367 |
| UN(4,3) | id | Species Virginica | 4.8824 | 2.2750 |
| UN(4,4) | id | Species Virginica | 7.5433 | 1.5240 |
| Tests of Covariance Parameters Based on the Restricted Likelihood |
|||||
|---|---|---|---|---|---|
| Label | DF | -2 Res Log Like | ChiSq | Pr > ChiSq | Note |
| Homogeneity | 20 | 2959.55 | 146.66 | <.0001 | DF |
Denote as  the covariance matrix for species
the covariance matrix for species 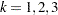 with elements
with elements 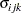 . In processing the COVTEST hypothesis
. In processing the COVTEST hypothesis 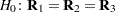 , the GLIMMIX procedure fits a model that satistfies the constraints
, the GLIMMIX procedure fits a model that satistfies the constraints
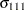 |
 |
|||
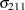 |
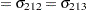 |
|||
 |
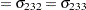 |
|||
 |
 |
|||
 |
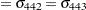 |
where 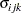 is the covariance between the
is the covariance between the 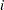 th and
th and 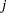 th variable for the
th variable for the 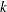 th species. The
th species. The  2 restricted log likelihood of this restricted model is 2959.55 (Output 40.9.4). The change of 146.66 compared to the full model is highly significant. There is sufficient evidence to reject the notion of equal covariance matrices among the three iris species.
2 restricted log likelihood of this restricted model is 2959.55 (Output 40.9.4). The change of 146.66 compared to the full model is highly significant. There is sufficient evidence to reject the notion of equal covariance matrices among the three iris species.
Equality of covariance matrices implies equality of correlation matrices, but the reverse is not true. Fewer constraints are needed to equate correlations because the diagonal entries of the covariance matrices are free to vary. In order to test the equality of the correlation matrices among the three species, you can parameterize the unstructured covariance matrix in terms of the correlations and use a COVTEST statement with general contrasts, as shown in the following statements:
ods select FitStatistics CovParms CovTests;
proc glimmix data=iris_univ;
class species var id;
model response = species*var;
random _residual_ / type=unr group=species subject=id;
covtest 'Equal Covariance Matrices' homogeneity;
covtest 'Equal Correlation Matrices' general
0 0 0 0 1 0 0 0 0 0
0 0 0 0 -1 0 0 0 0 0,
0 0 0 0 1 0 0 0 0 0
0 0 0 0 0 0 0 0 0 0
0 0 0 0 -1 0 0 0 0 0,
0 0 0 0 0 1 0 0 0 0
0 0 0 0 0 -1 0 0 0 0,
0 0 0 0 0 1 0 0 0 0
0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 -1 0 0 0 0,
0 0 0 0 0 0 1 0 0 0
0 0 0 0 0 0 -1 0 0 0,
0 0 0 0 0 0 1 0 0 0
0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 -1 0 0 0,
0 0 0 0 0 0 0 1 0 0
0 0 0 0 0 0 0 -1 0 0,
0 0 0 0 0 0 0 1 0 0
0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 -1 0 0,
0 0 0 0 0 0 0 0 1 0
0 0 0 0 0 0 0 0 -1 0,
0 0 0 0 0 0 0 0 1 0
0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 -1 0,
0 0 0 0 0 0 0 0 0 1
0 0 0 0 0 0 0 0 0 -1,
0 0 0 0 0 0 0 0 0 1
0 0 0 0 0 0 0 0 0 0
0 0 0 0 0 0 0 0 0 -1 / estimates;
run;
The TYPE=UNR structure is a reparameterization of TYPE=UN. The models provide the same fit, as seen by comparison of the "Fit Statistics" tables in Output 40.9.2 and Output 40.9.5. The covariance parameters are ordered differently, however. In each group, the four variances precede the six correlations (Output 40.9.5). The first COVTEST statement tests the homogeneity hypothesis in terms of the UNR parameterization, and the result is identical to the test in Output 40.9.4. The second COVTEST statement restricts the correlations to be equal across groups. If 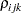 is the correlation between the
is the correlation between the 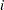 th and
th and 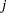 th variable for the
th variable for the 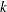 th species, the 12 restrictions are
th species, the 12 restrictions are
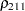 |
 |
|||
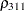 |
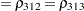 |
|||
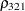 |
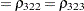 |
|||
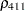 |
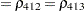 |
|||
 |
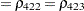 |
|||
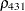 |
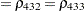 |
The ESTIMATES option in the COVTEST statement requests that the GLIMMIX procedure display the covariance parameter estimates in the restricted model (Output 40.9.5).
| Fit Statistics | |
|---|---|
| -2 Res Log Likelihood | 2812.89 |
| AIC (smaller is better) | 2872.89 |
| AICC (smaller is better) | 2876.23 |
| BIC (smaller is better) | 2963.21 |
| CAIC (smaller is better) | 2993.21 |
| HQIC (smaller is better) | 2909.58 |
| Generalized Chi-Square | 588.00 |
| Gener. Chi-Square / DF | 1.00 |
| Covariance Parameter Estimates | ||||
|---|---|---|---|---|
| Cov Parm | Subject | Group | Estimate | Standard Error |
| Var(1) | id | Species Setosa | 12.4249 | 2.5102 |
| Var(2) | id | Species Setosa | 14.3690 | 2.9030 |
| Var(3) | id | Species Setosa | 3.0159 | 0.6093 |
| Var(4) | id | Species Setosa | 1.1106 | 0.2244 |
| Corr(2,1) | id | Species Setosa | 0.7425 | 0.06409 |
| Corr(3,1) | id | Species Setosa | 0.2672 | 0.1327 |
| Corr(3,2) | id | Species Setosa | 0.1777 | 0.1383 |
| Corr(4,1) | id | Species Setosa | 0.2781 | 0.1318 |
| Corr(4,2) | id | Species Setosa | 0.2328 | 0.1351 |
| Corr(4,3) | id | Species Setosa | 0.3316 | 0.1271 |
| Var(1) | id | Species Versicolor | 26.6433 | 5.3828 |
| Var(2) | id | Species Versicolor | 9.8469 | 1.9894 |
| Var(3) | id | Species Versicolor | 22.0816 | 4.4612 |
| Var(4) | id | Species Versicolor | 3.9106 | 0.7901 |
| Corr(2,1) | id | Species Versicolor | 0.5259 | 0.1033 |
| Corr(3,1) | id | Species Versicolor | 0.7540 | 0.06163 |
| Corr(3,2) | id | Species Versicolor | 0.5605 | 0.09797 |
| Corr(4,1) | id | Species Versicolor | 0.5465 | 0.1002 |
| Corr(4,2) | id | Species Versicolor | 0.6640 | 0.07987 |
| Corr(4,3) | id | Species Versicolor | 0.7867 | 0.05445 |
| Var(1) | id | Species Virginica | 40.4343 | 8.1690 |
| Var(2) | id | Species Virginica | 10.4004 | 2.1012 |
| Var(3) | id | Species Virginica | 30.4588 | 6.1536 |
| Var(4) | id | Species Virginica | 7.5433 | 1.5240 |
| Corr(2,1) | id | Species Virginica | 0.4572 | 0.1130 |
| Corr(3,1) | id | Species Virginica | 0.8642 | 0.03616 |
| Corr(3,2) | id | Species Virginica | 0.4010 | 0.1199 |
| Corr(4,1) | id | Species Virginica | 0.2811 | 0.1316 |
| Corr(4,2) | id | Species Virginica | 0.5377 | 0.1015 |
| Corr(4,3) | id | Species Virginica | 0.3221 | 0.1280 |
| Tests of Covariance Parameters Based on the Restricted Likelihood |
|||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Label | DF | -2 Res Log Like | ChiSq | Pr > ChiSq | Estimates H0 | Note | |||||||||||||||||||||||||||||
| Est1 | Est2 | Est3 | Est4 | Est5 | Est6 | Est7 | Est8 | Est9 | Est10 | Est11 | Est12 | Est13 | Est14 | Est15 | Est16 | Est17 | Est18 | Est19 | Est20 | Est21 | Est22 | Est23 | Est24 | Est25 | Est26 | Est27 | Est28 | Est29 | Est30 | ||||||
| Equal Covariance Matrices | 20 | 2959.55 | 146.66 | <.0001 | 26.5004 | 11.5395 | 18.5179 | 4.1883 | 0.5302 | 0.7562 | 0.3779 | 0.3645 | 0.4705 | 0.4845 | 26.5004 | 11.5395 | 18.5179 | 4.1883 | 0.5302 | 0.7562 | 0.3779 | 0.3645 | 0.4705 | 0.4845 | 26.5004 | 11.5395 | 18.5179 | 4.1883 | 0.5302 | 0.7562 | 0.3779 | 0.3645 | 0.4705 | 0.4845 | DF |
| Equal Correlation Matrices | 12 | 2876.38 | 63.49 | <.0001 | 16.4715 | 14.8656 | 4.8427 | 1.4392 | 0.5612 | 0.6827 | 0.4016 | 0.3844 | 0.4976 | 0.5219 | 24.4020 | 9.1566 | 17.4434 | 3.0021 | 0.5612 | 0.6827 | 0.4016 | 0.3844 | 0.4976 | 0.5219 | 35.0544 | 10.8350 | 27.3593 | 8.1395 | 0.5612 | 0.6827 | 0.4016 | 0.3844 | 0.4976 | 0.5219 | DF |
The result of the homogeneity test is identical to that in Output 40.9.4. The hypothesis of equality of the correlation matrices is also rejected with a chi-square value of 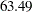 and a p-value of
and a p-value of  . Notice, however, that the chi-square statistic is smaller than in the test of homogeneity due to the smaller number of restrictions imposed on the full model. The estimate of the common correlation matrix in the restricted model is
. Notice, however, that the chi-square statistic is smaller than in the test of homogeneity due to the smaller number of restrictions imposed on the full model. The estimate of the common correlation matrix in the restricted model is
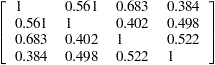 |