The GLIMMIX Procedure
-
Overview

-
Getting Started

-
Syntax
 PROC GLIMMIX Statement BY Statement CLASS Statement CONTRAST Statement COVTEST Statement EFFECT Statement ESTIMATE Statement FREQ Statement ID Statement LSMEANS Statement LSMESTIMATE Statement MODEL Statement NLOPTIONS Statement OUTPUT Statement PARMS Statement RANDOM Statement SLICE Statement STORE Statement WEIGHT Statement Programming Statements User-Defined Link or Variance Function
PROC GLIMMIX Statement BY Statement CLASS Statement CONTRAST Statement COVTEST Statement EFFECT Statement ESTIMATE Statement FREQ Statement ID Statement LSMEANS Statement LSMESTIMATE Statement MODEL Statement NLOPTIONS Statement OUTPUT Statement PARMS Statement RANDOM Statement SLICE Statement STORE Statement WEIGHT Statement Programming Statements User-Defined Link or Variance Function -
Details
 Generalized Linear Models Theory Generalized Linear Mixed Models Theory GLM Mode or GLMM Mode Statistical Inference for Covariance Parameters Satterthwaite Degrees of Freedom Approximation Empirical Covariance (Sandwich) Estimators Exploring and Comparing Covariance Matrices Processing by Subjects Radial Smoothing Based on Mixed Models Odds and Odds Ratio Estimation Parameterization of Generalized Linear Mixed Models Response-Level Ordering and Referencing Comparing the GLIMMIX and MIXED Procedures Singly or Doubly Iterative Fitting Default Estimation Techniques Default Output Notes on Output Statistics ODS Table Names ODS Graphics
Generalized Linear Models Theory Generalized Linear Mixed Models Theory GLM Mode or GLMM Mode Statistical Inference for Covariance Parameters Satterthwaite Degrees of Freedom Approximation Empirical Covariance (Sandwich) Estimators Exploring and Comparing Covariance Matrices Processing by Subjects Radial Smoothing Based on Mixed Models Odds and Odds Ratio Estimation Parameterization of Generalized Linear Mixed Models Response-Level Ordering and Referencing Comparing the GLIMMIX and MIXED Procedures Singly or Doubly Iterative Fitting Default Estimation Techniques Default Output Notes on Output Statistics ODS Table Names ODS Graphics -
Examples
 Binomial Counts in Randomized Blocks Mating Experiment with Crossed Random Effects Smoothing Disease Rates; Standardized Mortality Ratios Quasi-likelihood Estimation for Proportions with Unknown Distribution Joint Modeling of Binary and Count Data Radial Smoothing of Repeated Measures Data Isotonic Contrasts for Ordered Alternatives Adjusted Covariance Matrices of Fixed Effects Testing Equality of Covariance and Correlation Matrices Multiple Trends Correspond to Multiple Extrema in Profile Likelihoods Maximum Likelihood in Proportional Odds Model with Random Effects Fitting a Marginal (GEE-Type) Model Response Surface Comparisons with Multiplicity Adjustments Generalized Poisson Mixed Model for Overdispersed Count Data Comparing Multiple B-Splines Diallel Experiment with Multimember Random Effects Linear Inference Based on Summary Data
Binomial Counts in Randomized Blocks Mating Experiment with Crossed Random Effects Smoothing Disease Rates; Standardized Mortality Ratios Quasi-likelihood Estimation for Proportions with Unknown Distribution Joint Modeling of Binary and Count Data Radial Smoothing of Repeated Measures Data Isotonic Contrasts for Ordered Alternatives Adjusted Covariance Matrices of Fixed Effects Testing Equality of Covariance and Correlation Matrices Multiple Trends Correspond to Multiple Extrema in Profile Likelihoods Maximum Likelihood in Proportional Odds Model with Random Effects Fitting a Marginal (GEE-Type) Model Response Surface Comparisons with Multiplicity Adjustments Generalized Poisson Mixed Model for Overdispersed Count Data Comparing Multiple B-Splines Diallel Experiment with Multimember Random Effects Linear Inference Based on Summary Data - References
| ODS Table Names |
Each table created by PROC GLIMMIX has a name associated with it, and you must use this name to reference the table when you use ODS statements. These names are listed in Table 40.20.
Table Name |
Description |
Required Statement / Option |
|---|---|---|
AsyCorr |
asymptotic correlation matrix of covariance parameters |
PROC GLIMMIX ASYCORR |
AsyCov |
asymptotic covariance matrix of covariance parameters |
PROC GLIMMIX ASYCOV |
CholG |
Cholesky root of the estimated |
|
CholV |
Cholesky root of blocks of the estimated |
|
ClassLevels |
level information from the CLASS statement |
default output |
Coef |
|
E option in MODEL, CONTRAST, ESTIMATE, LSMESTIMATE, or LSMEANS; ELSM option in LSMESTIMATE |
ColumnNames |
name association for OUTDESIGN data set |
PROC GLIMMIX |
CondFitStatistics |
conditional fit statistics |
|
Contrasts |
results from the CONTRAST statements |
|
ConvergenceStatus |
status of optimization at conclusion |
default output |
CorrB |
approximate correlation matrix of fixed-effects parameter estimates |
|
CovB |
approximate covariance matrix of fixed-effects parameter estimates |
|
CovBDetails |
details about model-based and/or adjusted covariance matrix of fixed effects |
|
CovBI |
inverse of approximate covariance matrix of fixed-effects parameter estimates |
|
CovBModelBased |
model-based (unadjusted) covariance matrix of fixed effects if DDFM=KR or EMPIRICAL option is used |
|
CovParms |
estimated covariance parameters in GLMMs |
default output (in GLMMs) |
CovTests |
results from COVTEST statements (except for confidence bounds) |
|
Diffs |
differences of LS-means |
|
Dimensions |
dimensions of the model |
default output |
Estimates |
results from ESTIMATE statements |
|
FitStatistics |
fit statistics |
default |
G |
estimated |
|
GCorr |
correlation matrix from the estimated |
|
Hessian |
Hessian matrix (observed or expected) |
PROC GLIMMIX HESSIAN |
InvCholG |
inverse Cholesky root of the estimated |
|
InvCholV |
inverse Cholesky root of the blocks of the estimated |
|
InvG |
inverse of the estimated |
|
InvV |
inverse of blocks of the estimated |
|
IterHistory |
iteration history |
default output |
kdTree |
k-d tree information |
RANDOM / TYPE=RSMOOTH KNOTMETHOD= KDTREE(TREEINFO) |
KnotInfo |
knot coordinates of low-rank spline smoother |
|
LSMeans |
LS-means |
|
LSMEstimates |
estimates among LS-means |
|
LSMFtest |
F test for LSMESTIMATEs |
|
LSMLines |
lines display for LS-means |
|
ModelInfo |
model information |
default output |
NObs |
number of observations read and used, number of trials and events |
default output |
OddsRatios |
odds ratios of parameter estimates |
|
OptInfo |
optimization information |
default output |
ParameterEstimates |
fixed-effects solution; overdispersion and scale parameter in GLMs |
|
ParmSearch |
parameter search values |
|
QuadCheck |
adaptive recalculation of quadrature approximation at solution |
|
ResponseProfile |
response categories and category modeled |
default output in models with binary or nominal response |
Slices |
tests of LS-means slices |
|
SliceDiffs |
differences of simple LS-means effects |
|
SolutionR |
random-effects solution vector |
|
StandardizedCoefficients |
fixed-effects solutions from centered and/or scaled model |
|
Tests1 |
Type I tests of fixed effects |
|
Tests2 |
Type II tests of fixed effects |
|
Tests3 |
Type III tests of fixed effects |
default output |
V |
blocks of the estimated |
|
VCorr |
correlation matrix from the blocks of the estimated |
The SLICE statement also creates tables, which are not listed in Table 40.20. For information about these tables, see the section SLICE Statement of Chapter 19, Shared Concepts and Topics.
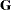 matrix
matrix 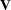 matrix
matrix 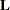 matrix coefficients
matrix coefficients