The UNIVARIATE Procedure
- Overview
-
Getting Started

-
Syntax

-
Details
 Missing ValuesRoundingDescriptive StatisticsCalculating the ModeCalculating PercentilesTests for LocationConfidence Limits for Parameters of the Normal DistributionRobust EstimatorsCreating Line Printer PlotsCreating High-Resolution GraphicsUsing the CLASS Statement to Create Comparative PlotsPositioning InsetsFormulas for Fitted Continuous DistributionsGoodness-of-Fit TestsKernel Density EstimatesConstruction of Quantile-Quantile and Probability PlotsInterpretation of Quantile-Quantile and Probability PlotsDistributions for Probability and Q-Q PlotsEstimating Shape Parameters Using Q-Q PlotsEstimating Location and Scale Parameters Using Q-Q PlotsEstimating Percentiles Using Q-Q PlotsInput Data SetsOUT= Output Data Set in the OUTPUT StatementOUTHISTOGRAM= Output Data SetOUTKERNEL= Output Data SetOUTTABLE= Output Data SetTables for Summary StatisticsODS Table NamesODS Tables for Fitted DistributionsODS GraphicsComputational Resources
Missing ValuesRoundingDescriptive StatisticsCalculating the ModeCalculating PercentilesTests for LocationConfidence Limits for Parameters of the Normal DistributionRobust EstimatorsCreating Line Printer PlotsCreating High-Resolution GraphicsUsing the CLASS Statement to Create Comparative PlotsPositioning InsetsFormulas for Fitted Continuous DistributionsGoodness-of-Fit TestsKernel Density EstimatesConstruction of Quantile-Quantile and Probability PlotsInterpretation of Quantile-Quantile and Probability PlotsDistributions for Probability and Q-Q PlotsEstimating Shape Parameters Using Q-Q PlotsEstimating Location and Scale Parameters Using Q-Q PlotsEstimating Percentiles Using Q-Q PlotsInput Data SetsOUT= Output Data Set in the OUTPUT StatementOUTHISTOGRAM= Output Data SetOUTKERNEL= Output Data SetOUTTABLE= Output Data SetTables for Summary StatisticsODS Table NamesODS Tables for Fitted DistributionsODS GraphicsComputational Resources -
Examples
 Computing Descriptive Statistics for Multiple VariablesCalculating ModesIdentifying Extreme Observations and Extreme ValuesCreating a Frequency TableCreating Basic Summary PlotsAnalyzing a Data Set With a FREQ VariableSaving Summary Statistics in an OUT= Output Data SetSaving Percentiles in an Output Data SetComputing Confidence Limits for the Mean, Standard Deviation, and VarianceComputing Confidence Limits for Quantiles and PercentilesComputing Robust EstimatesTesting for LocationPerforming a Sign Test Using Paired DataCreating a HistogramCreating a One-Way Comparative HistogramCreating a Two-Way Comparative HistogramAdding Insets with Descriptive StatisticsBinning a HistogramAdding a Normal Curve to a HistogramAdding Fitted Normal Curves to a Comparative HistogramFitting a Beta CurveFitting Lognormal, Weibull, and Gamma CurvesComputing Kernel Density EstimatesFitting a Three-Parameter Lognormal CurveAnnotating a Folded Normal CurveCreating Lognormal Probability PlotsCreating a Histogram to Display Lognormal FitCreating a Normal Quantile PlotAdding a Distribution Reference LineInterpreting a Normal Quantile PlotEstimating Three Parameters from Lognormal Quantile PlotsEstimating Percentiles from Lognormal Quantile PlotsEstimating Parameters from Lognormal Quantile PlotsComparing Weibull Quantile PlotsCreating a Cumulative Distribution PlotCreating a P-P Plot
Computing Descriptive Statistics for Multiple VariablesCalculating ModesIdentifying Extreme Observations and Extreme ValuesCreating a Frequency TableCreating Basic Summary PlotsAnalyzing a Data Set With a FREQ VariableSaving Summary Statistics in an OUT= Output Data SetSaving Percentiles in an Output Data SetComputing Confidence Limits for the Mean, Standard Deviation, and VarianceComputing Confidence Limits for Quantiles and PercentilesComputing Robust EstimatesTesting for LocationPerforming a Sign Test Using Paired DataCreating a HistogramCreating a One-Way Comparative HistogramCreating a Two-Way Comparative HistogramAdding Insets with Descriptive StatisticsBinning a HistogramAdding a Normal Curve to a HistogramAdding Fitted Normal Curves to a Comparative HistogramFitting a Beta CurveFitting Lognormal, Weibull, and Gamma CurvesComputing Kernel Density EstimatesFitting a Three-Parameter Lognormal CurveAnnotating a Folded Normal CurveCreating Lognormal Probability PlotsCreating a Histogram to Display Lognormal FitCreating a Normal Quantile PlotAdding a Distribution Reference LineInterpreting a Normal Quantile PlotEstimating Three Parameters from Lognormal Quantile PlotsEstimating Percentiles from Lognormal Quantile PlotsEstimating Parameters from Lognormal Quantile PlotsComparing Weibull Quantile PlotsCreating a Cumulative Distribution PlotCreating a P-P Plot - References
Kernel Density Estimates
You can use the KERNEL option to superimpose kernel density estimates on histograms. Smoothing the data distribution with a kernel density estimate can be more effective than using a histogram to identify features that might be obscured by the choice of histogram bins or sampling variation. A kernel density estimate can also be more effective than a parametric curve fit when the process distribution is multi-modal. See Example 4.23.
The general form of the kernel density estimator is
![\[ \hat{f}_{\lambda }(x) = \frac{hv}{n\lambda } \sum ^ n_{i=1}K_{0}\left(\frac{x-x_{i}}{\lambda }\right) \]](images/procstat_univariate0403.png)
where
-
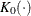 is the kernel function
is the kernel function
-
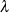 is the bandwidth
is the bandwidth
-
n is the sample size
-
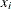 is the ith observation
is the ith observation
-
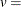 vertical scaling factor
vertical scaling factor
and
![\[ v = \left\{ \begin{array}{ll} n & \mbox{for VSCALE=COUNT} \\ 100 & \mbox{for VSCALE=PERCENT} \\ 1 & \mbox{for VSCALE=PROPORTION} \end{array} \right. \]](images/procstat_univariate0405.png)
The KERNEL option provides three kernel functions (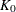 ): normal, quadratic, and triangular. You can specify the function with the K= kernel-option in parentheses after the KERNEL option. Values for the K= option are NORMAL, QUADRATIC, and TRIANGULAR (with aliases of N,
Q, and T, respectively). By default, a normal kernel is used. The formulas for the kernel functions are
): normal, quadratic, and triangular. You can specify the function with the K= kernel-option in parentheses after the KERNEL option. Values for the K= option are NORMAL, QUADRATIC, and TRIANGULAR (with aliases of N,
Q, and T, respectively). By default, a normal kernel is used. The formulas for the kernel functions are
![\[ \begin{array}{lll} \mbox{Normal} & K_0(t) = \frac{1}{\sqrt {2\pi }} \exp (-\frac{1}{2}t^{2}) & \mbox{for } -\infty < t < \infty \\ \mbox{Quadratic} & K_0(t) = \frac{3}{4}(1-t^2) & \mbox{for } |t| \leq 1 \\ \mbox{Triangular} & K_0(t) = 1-|t| & \mbox{for } |t| \leq 1 \end{array} \]](images/procstat_univariate0407.png)
The value of 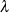 , referred to as the bandwidth parameter, determines the degree of smoothness in the estimated density function. You specify
, referred to as the bandwidth parameter, determines the degree of smoothness in the estimated density function. You specify
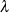 indirectly by specifying a standardized bandwidth c with the C= kernel-option. If Q is the interquartile range and n is the sample size, then c is related to
indirectly by specifying a standardized bandwidth c with the C= kernel-option. If Q is the interquartile range and n is the sample size, then c is related to 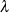 by the formula
by the formula
![\[ \lambda = cQn^{-\frac{1}{5}} \]](images/procstat_univariate0408.png)
For a specific kernel function, the discrepancy between the density estimator 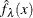 and the true density
and the true density 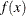 is measured by the mean integrated square error (MISE):
is measured by the mean integrated square error (MISE):
![\[ \mbox{MISE}(\lambda ) = \int _{x}\{ E(\hat{f}_{\lambda }(x)) - f(x)\} ^{2}dx + \int _{x}var(\hat{f}_{\lambda }(x))dx \]](images/procstat_univariate0411.png)
The MISE is the sum of the integrated squared bias and the variance. An approximate mean integrated square error (AMISE) is:
![\[ \mbox{AMISE}(\lambda ) = \frac{1}{4}\lambda ^{4} \left(\int _{t}t^{2}K(t)dt\right)^2 \int _ x\left(f^{\prime \prime }(x)\right)^2dx + \frac{1}{n\lambda }\int _{t}K(t)^2dt \]](images/procstat_univariate0412.png)
A bandwidth that minimizes AMISE can be derived by treating 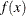 as the normal density that has parameters
as the normal density that has parameters 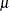 and
and  estimated by the sample mean and standard deviation. If you do not specify a bandwidth parameter or if you specify C=MISE,
the bandwidth that minimizes AMISE is used. The value of AMISE can be used to compare different density estimates. You can
also specify C=SJPI to select the bandwidth by using a plug-in formula of Sheather and Jones (Jones, Marron, and Sheather
1996). For each estimate, the bandwidth parameter c, the kernel function type, and the value of AMISE are reported in the SAS log.
estimated by the sample mean and standard deviation. If you do not specify a bandwidth parameter or if you specify C=MISE,
the bandwidth that minimizes AMISE is used. The value of AMISE can be used to compare different density estimates. You can
also specify C=SJPI to select the bandwidth by using a plug-in formula of Sheather and Jones (Jones, Marron, and Sheather
1996). For each estimate, the bandwidth parameter c, the kernel function type, and the value of AMISE are reported in the SAS log.
The general kernel density estimates assume that the domain of the density to estimate can take on all values on a real line.
However, sometimes the domain of a density is an interval bounded on one or both sides. For example, if a variable Y is a measurement of only positive values, then the kernel density curve should be bounded so that is zero for negative Y values. You can use the LOWER= and UPPER= kernel-options to specify the bounds.
The UNIVARIATE procedure uses a reflection technique to create the bounded kernel density curve, as described in Silverman
(1986, pp. 30-31). It adds the reflections of the kernel density that are outside the boundary to the bounded kernel estimates.
The general form of the bounded kernel density estimator is computed by replacing 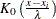 in the original equation with
in the original equation with
![\[ \left\{ K_0\left(\frac{x - x_ i}{\lambda }\right) + K_0\left(\frac{(x - x_ l) + (x_ i - x_ l)}{\lambda }\right) + K_0\left(\frac{(x_ u - x) + (x_ u - x_ i)}{\lambda }\right) \right\} \]](images/procstat_univariate0414.png)
where 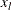 is the lower bound and
is the lower bound and 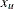 is the upper bound.
is the upper bound.
Without a lower bound, 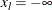 and
and 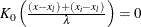 . Similarly, without an upper bound,
. Similarly, without an upper bound, 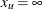 and
and 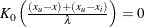 .
.
When C=MISE is used with a bounded kernel density, the UNIVARIATE procedure uses a bandwidth that minimizes the AMISE for its corresponding unbounded kernel.