-
ALPHA=number
-
specifies that a confidence level of 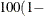 number
number 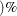 is to be used in forming bootstrap confidence intervals for the penetrance parameters when the BOOT= option is given. This
value of number must be between 0 and 1 and is set to 0.05 by default.
is to be used in forming bootstrap confidence intervals for the penetrance parameters when the BOOT= option is given. This
value of number must be between 0 and 1 and is set to 0.05 by default.
-
BOOT=number
-
requests that confidence intervals be calculated for the penetrance parameters by using number iterations of the bootstrap. You must input which recombination parameters ( s) to use in the calculation by using the R option or an error is generated.
s) to use in the calculation by using the R option or an error is generated.
-
CROSS=BACK | B | DH
CROSS=INTER | F
-
specifies the type of cross for the input data set. The options include BACK or B for a backcross or, equivalently, DH for
a doubled-haploid population. The other option is INTER or F for an F intercross. The default is backcross.
-
GEN=number
-
specifies the generation number of the offspring in the input data set. Valid values include any integer greater than or equal
to one. The default is 1.
-
HETEROZYGOTE=“heterozygote”
HE=“heterozygote”
-
specifies the value for the heterozygous genotype used in the input data set. The default value is “B.”
-
HOMOZYGOTE=“homozygote”
HO=“homozygote”
-
specifies the value for the homozygous genotype used in the input data set. The default value is “A.” If the experimental design is an F cross, then this is the genotype homozygous for the parent 1 allele.
-
HOMOZYGOTE2=“homozygote2”
HO2=“homozygote2”
-
specifies the value for the genotype homozygous in the parent 2 allele used in the input data set. The default value is “C.”
-
LINKMOD=HALDANE | H
LINKMOD=KOSAMBI | K
-
specifies the model to be used to calculate the marker recombination parameters from the marker location values in the map
data set. The options include Haldane and Kosambi. The default value is Haldane.
-
LINKUNIT=CM | C
LINKUNIT=RECDIST | R
-
specifies the units used for the marker location variable in the marker data set. The options include centimorgans or recombination
distance (kilobases). The default value is centimorgans.
-
OUTSTAT=SAS-data-set
-
names the SAS data set to be used for the parameter estimates and, when the BOOT= option is specified, confidence intervals.
-
PMAX=number
-
specifies the highest penetrance value that is considered in range and included in the output. Any real number is a valid
value as long as it is greater than PMIN. The PMAX option is ignored if the R option, which precludes a grid search, is used.
By default, there is no upper limit for the range of penetrance values included in the output.
-
PMIN=number
-
specifies the lowest penetrance value that is considered in range and included in the output. Any real number is a valid value
as long as it is less than PMAX. The PMIN option is ignored if the R option, which precludes a grid search, is used. By default,
there is no lower limit for the range of penetrance values included in the output.
-
R=number-list
-
specifies the values of  (recombination parameters) used to estimate the penetrance parameters. There is one
(recombination parameters) used to estimate the penetrance parameters. There is one  for each of the
for each of the 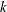 adjacent marker/BTL pairs, where
adjacent marker/BTL pairs, where 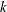 is the number of markers in the MARKER statement. A list of values can be given to specify a different
is the number of markers in the MARKER statement. A list of values can be given to specify a different  for each pair, or a single value can be specified to be used for all
for each pair, or a single value can be specified to be used for all  . If there are fewer than
. If there are fewer than 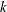 values specified, the last value given is used for the remaining
values specified, the last value given is used for the remaining  . If the R option is used to specify
. If the R option is used to specify  , the grid search parameters (RSTART, REND, and RINC) are ignored. The R option is required if the BOOT= option is specified.
These
, the grid search parameters (RSTART, REND, and RINC) are ignored. The R option is required if the BOOT= option is specified.
These  s are used to calculate the confidence intervals of the penetrance parameters in the bootstrap calculation. Each
s are used to calculate the confidence intervals of the penetrance parameters in the bootstrap calculation. Each  must be a real number greater than or equal to 0 and less than 0.5, and invalid values are replaced by the default value
of 0.
must be a real number greater than or equal to 0 and less than 0.5, and invalid values are replaced by the default value
of 0.
-
REND=number
-
specifies the ending value for each recombination parameter in the grid search. The default value is 0.5. REND must be a real
number greater than 0 and less than or equal to 0.5.
-
RINC=number
-
specifies the increment to be used for the recombination parameter grid search. The default value is 0.1. Any real number
greater than 0 and less than or equal to 0.5 is valid.
-
RSTART=number
-
specifies the starting value for each recombination parameter in the grid search. The default value is 0. RSTART must be a
real number from 0 to (but not including) 0.5.
-
SEED=number
-
specifies the initial seed for the random number generator used for creating the bootstrap samples when the BOOT= option is
given. The value for number must be an integer; the computer clock time is used if the option is omitted or the integer specified is less than or equal
to 0. For more details about seed values, see
SAS Language Reference: Concepts.
-
THETA=number-list
-
specifies the values of recombination probabilities between adjacent pairs of markers listed in the MARKER statement. There
is one 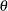 for each of the
for each of the 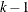 pairs of adjacent markers, where
pairs of adjacent markers, where 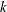 is the number of markers specified in the MARKER statement. A list of values can be given to specify a different
is the number of markers specified in the MARKER statement. A list of values can be given to specify a different 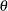 for each pair, or a single value can be specified to be used for all
for each pair, or a single value can be specified to be used for all 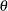 s. If there are fewer than
s. If there are fewer than 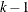 values specified, the last value given is used for the remaining
values specified, the last value given is used for the remaining 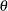 .
Note: If the MAP= data set is specified and contains the variable
.
Note: If the MAP= data set is specified and contains the variable Location, the 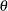 values are calculated using these distances and this option is ignored. If locations are not provided in the MAP= data set
and this option is omitted, then default values of
values are calculated using these distances and this option is ignored. If locations are not provided in the MAP= data set
and this option is omitted, then default values of 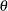 of 0.5 are used. Each
of 0.5 are used. Each 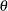 must be a real number between 0 and 0.5, and invalid values are replaced by the default value of 0.5.
must be a real number between 0 and 0.5, and invalid values are replaced by the default value of 0.5.
![]() s are displayed. Alternatively, a specific set of
s are displayed. Alternatively, a specific set of ![]() values can be specified using the R option.
values can be specified using the R option.
![]() performed for a model with several markers (
performed for a model with several markers (![]() ) can be lengthy and computationally intensive. The computational time increases as
) can be lengthy and computationally intensive. The computational time increases as ![]() increases, where
increases, where ![]() is the number of increments of
is the number of increments of ![]() and
and ![]() is the number of markers. For this reason, it is recommended that a model with several markers use consecutive grid searches
with very few increments (small
is the number of markers. For this reason, it is recommended that a model with several markers use consecutive grid searches
with very few increments (small ![]() ) in order to zero in on the correct values of
) in order to zero in on the correct values of ![]() .
.
![]() parameters are calculated from the map data set by using whichever model is specified in the LINKMOD= option (Haldane is
the default). Alternatively, a specific set of
parameters are calculated from the map data set by using whichever model is specified in the LINKMOD= option (Haldane is
the default). Alternatively, a specific set of ![]() values can be specified using the THETA option. A confidence interval of significance level
values can be specified using the THETA option. A confidence interval of significance level ![]() can be requested by using the BOOT= option and specifying the number of bootstrap iterations.
can be requested by using the BOOT= option and specifying the number of bootstrap iterations.
