The BTL Procedure (Experimental)
This section describes the displayed output from PROC BTL. See the section ODS Table Names for details about how this output interfaces with the Output Delivery System.
The “Model Statistics” table is displayed by default and contains the following information about the fitted model:
-
LinkageGroup, the linkage group of a marker (displayed for single marker effects only if the GROUP= option is given in the MARKER statement)
-
MarkerEffect, the marker(s) constituting the fixed marker effect
-
Name, the name of the marker (displayed for single marker effects only if a
Namevariable is contained in the MAP= data set) -
DF, the degrees of freedom for the chi-squared test statistic
-
ChiSquare, the test statistic for the likelihood ratio of the full model to the null model
-
ProbChiSq, the
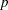 -value of the chi-squared test statistic
-value of the chi-squared test statistic
-
AIC, Akaike’s information criterion (Akaike 1974)
-
AICC, a finite-sample corrected version of AIC (Burnham and Anderson 1998)
-
BIC, Bayesian information criterion (Schwarz 1978)
The “Marker Class Means” table is displayed by default if the PARMEST statement is used and contains the average binary trait for each marker class. It contains the following:
-
Parameter, the marker class parameter
-
MarkerGenotype, the marker class genotype
-
N, the number of individuals in the marker class
-
Mean, the MLE of the marker class mean, which is the proportion of individuals in the marker class with trait 1
-
StdErr, the standard error of the marker class mean estimate
The “Parameter Estimates” table is displayed by default if the PARMEST statement is used and contains the estimated recombination and penetrance parameters
for the estimated BTL model. It contains the model number, the parameter name, the parameter estimate, and the ![]() confidence interval for the estimate if the BOOT= option is used.
confidence interval for the estimate if the BOOT= option is used.
