The ALLELE Procedure
PROC ALLELE <options> ;
You can specify the following options in the PROC ALLELE statement.
-
ALLELEMIN=number
AMIN=number -
indicates that only alleles with a frequency greater than or equal to number should be included in the “Allele Frequencies” table. By default, any allele that appears in a nonmissing genotype in the sample is included in the table. The value of number must be between 0 and 1.
- ALPHA=number
-
specifies that a confidence level of
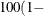 number
number 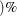 is to be used in forming bootstrap confidence intervals for estimates of allele frequencies and disequilibrium coefficients.
The value of number must be between 0 and 1, and is set to 0.05 by default.
is to be used in forming bootstrap confidence intervals for estimates of allele frequencies and disequilibrium coefficients.
The value of number must be between 0 and 1, and is set to 0.05 by default.
-
BOOTSTRAP=number
BOOT=number -
indicates that bootstrap confidence intervals should be formed for the estimates of allele frequencies and one-locus disequilibrium coefficients by using number random samples. One thousand samples are usually recommended to form confidence intervals. If this statement is omitted, no confidence limits are reported.
- CORRCOEFF
-
requests that the “Linkage Disequilibrium Measures” table be displayed and contain the correlation coefficient
 , a linkage disequilibrium measure.
, a linkage disequilibrium measure.
- DATA=SAS-data-set
-
names the input SAS data set to be used by PROC ALLELE. The default is to use the most recently created data set.
- DELIMITER=’string’
-
indicates the string that is used to separate the two alleles that compose the genotypes contained in the variables specified in the VAR statement. This option is ignored if GENOCOL is not specified.
- DELTA
-
requests that the “Linkage Disequilibrium Measures” table be displayed and contain the population attributable risk
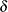 , a linkage disequilibrium measure. This option is ignored if HAPLO=NONE or NONEHWD.
, a linkage disequilibrium measure. This option is ignored if HAPLO=NONE or NONEHWD.
- DPRIME
-
requests that the “Linkage Disequilibrium Measures” table be displayed and contain Lewontin’s
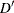 , a linkage disequilibrium measure.
, a linkage disequilibrium measure.
- GENOCOL
-
indicates that columns specified in the VAR statement contain genotypes instead of alleles. When this option is specified, there is one column per marker. The genotypes must consist of the two alleles separated by a delimiter. For a genotype with one missing allele, use a blank space to indicate a missing value; if both alleles are missing, either use a single missing value for the entire genotype or use the delimiter alone.
-
GENOMIN=number
GMIN=number -
indicates that only genotypes with a frequency greater than or equal to number should be included in the “Genotype Frequencies” table. By default, any genotype that appears at least once in the sample is included in the table. The value of number must be between 0 and 1.
-
HAPLO=NONE
HAPLO=EST
HAPLO=GIVEN
HAPLO=NONEHWD -
indicates whether haplotype frequencies should not be used, haplotype frequencies should be estimated, or observed haplotype frequencies in the data should be used. This option affects all linkage disequilibrium tests and measures. By default or when HAPLO=NONE or NONEHWD is specified, the composite linkage disequilibrium (CLD) coefficient is used in place of the usual linkage disequilibrium (LD) coefficient. In addition, the composite haplotype frequencies are used to form the linkage disequilibrium measures indicated by the options CORRCOEFF and DPRIME. When HAPLO=EST, the maximum likelihood estimates of the haplotype frequencies are used to calculate the LD test statistic as well as the LD measures. The HAPLO=GIVEN option indicates that the haplotypes have been observed, and thus the observed haplotype frequencies are used in the LD test statistic and measures.
When HAPLO=GIVEN, haplotypes are denoted in the data in the following manner according to the type of input data used:
-
If you omit the TALL option in the PROC ALLELE statement, then all alleles contained in one of an individual’s two haplotypes must be in the first of the two variables listed for each marker, and alleles of the other haplotype must be in the second of the two variables listed for each marker. Similarly, if the GENOCOL option is used, the alleles composing one haplotype should all be the first allele listed in each genotype, and alleles of the other haplotype should be listed second.
-
If you specify the TALL option, then the alleles that compose one haplotype for an individual must all be in the first variable in the VAR statement, and all the alleles in the other haplotype must be in the second variable in the VAR statement. When the GENOCOL option is also specified, the alleles of one haplotype should all be the first allele listed in the genotype, and alleles of the other haplotype should be listed second.
-
-
HAPLOMIN=number
HMIN=number -
indicates that only haplotypes with a frequency greater than or equal to number should be included in the “Linkage Disequilibrium Measures” table. By default, any haplotype that appears in the sample (or is estimated to appear at least once) is included in the table. The value of number must be between 0 and 1.
-
INDIVIDUAL=variable
INDIV=variable -
specifies the individual ID variable when using the TALL option. This variable can be character or numeric.
- LOGNOTE
-
requests that notes be written to the log indicating the status of the LD calculations.
- MARKER=variable
-
specifies the marker ID variable when using the TALL option. This variable contains the names of the markers that are used in all output and can be character or numeric.
- MAXDIST=number
-
specifies the maximum distance possible between a pair of markers in order for linkage disequilibrium calculations to be performed on that pair. If the NDATA= option is not specified or the NDATA= data set does not contain a
LOCATIONvariable, then the distance between a pair of markers is the number of markers apart that they are, assuming that markers are specified in the VAR statement in the physical order in which they appear on a chromosome. For example, if MAXDIST=1 is specified, linkage disequilibrium measures and statistics are calculated only for pairs of markers that are one marker apart, such as M1 and M2, M2 and M3, and so on. If there is an NDATA= data set specified that contains aLOCATIONvariable, then distances between markers are calculated as differences between values of this variable. Note that markers with missing values for theLOCATIONvariable are paired with all other markers for LD calculations. The default value is MAXDIST=50. - NDATA=SAS-data-set
-
names the input SAS data set containing names, or identifiers, for the markers used in the output. There must be a
NAMEvariable in this data set, which should contain the same number of rows as there are markers in the input data set specified in the DATA= option. When there are fewer rows than there are markers, markers without a name are named using the PREFIX= option. Likewise, if there is no NDATA= data set specified, the PREFIX= option is used. Note that this data set is ignored if the TALL option is specified in the PROC ALLELE statement. In that case, the marker variable names are taken from the marker ID variable specified in the MARKER= option.If there is a
LOCATIONvariable in the NDATA= data set, the value given for the MAXDIST= option for LD measures and testing is applied to distances between markers based on values of this variable. - NOFREQ
-
suppresses the display of the “Allele Frequencies” and the “Genotype Frequencies” tables.
See the section Displayed Output for a detailed description of these tables.
- NOPRINT
-
suppresses the normal display of results. Note that this option temporarily disables the Output Delivery System (ODS).
- OUTSTAT=SAS-data-set
-
names the output SAS data set containing the disequilibrium statistics, for both within-marker and between-marker disequilibria.
-
PERMS=number
EXACT=number -
indicates that Monte Carlo estimates of the exact
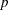 -values for the disequilibrium tests should be calculated using number permutations. Large values of number (10,000 or more) are usually recommended for accuracy, but long execution times can result, particularly with large data
sets. When this option is omitted, no permutations are performed and asymptotic
-values for the disequilibrium tests should be calculated using number permutations. Large values of number (10,000 or more) are usually recommended for accuracy, but long execution times can result, particularly with large data
sets. When this option is omitted, no permutations are performed and asymptotic 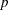 -values are reported. If HAPLO=EST, then only the exact tests for Hardy-Weinberg equilibrium are performed; the exact tests
for linkage disequilibrium cannot be performed since haplotypes are unknown.
-values are reported. If HAPLO=EST, then only the exact tests for Hardy-Weinberg equilibrium are performed; the exact tests
for linkage disequilibrium cannot be performed since haplotypes are unknown.
- PREFIX=prefix
-
specifies a prefix to use in constructing names for marker variables in all output. For example, if PREFIX=VAR, the names of the variables are VAR1, VAR2, …, VARn. Note that this option is ignored when the NDATA= option is specified, unless there are fewer names in the NDATA data set than there are markers; it is also ignored if the TALL option is specified, in which case the marker variable names are taken from the marker ID variable specified in the MARKER= option. Otherwise, if this option is omitted, PREFIX=M is the default when variables contain alleles; if GENOCOL is specified, then the names of the variables specified in the VAR statement are used as the marker names.
- PROPDIFF
-
requests that the “Linkage Disequilibrium Measures” table be displayed and contain the proportional difference
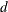 , a linkage disequilibrium measure. This option is ignored if HAPLO=NONE or NONEHWD.
, a linkage disequilibrium measure. This option is ignored if HAPLO=NONE or NONEHWD.
- RHO
-
requests that the “Linkage Disequilibrium Measures” table be displayed and contain
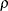 , a linkage disequilibrium measure, and its information
, a linkage disequilibrium measure, and its information 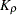 . This option is ignored if HAPLO=NONE or NONEHWD.
. This option is ignored if HAPLO=NONE or NONEHWD.
- SEED=number
-
specifies the initial seed for the random number generator used for permuting the data in the exact tests and for the bootstrap samples. The value for number must be an integer; the computer clock time is used if the option is omitted or the integer specified is less than or equal to 0. For more details about seed values, see SAS Language Reference: Concepts.
- TALL
-
indicates that the input data set is of an alternative format. This format contains the following columns: two containing marker alleles (or one containing marker genotypes if GENOCOL is specified), one for the marker identifier, and one for the individual identifier. The MARKER= and INDIV= options must also be specified for this option to be in effect. Note that when this option is used, the DATA= data set must first be sorted by any BY variables, then sorted by the marker ID variable, and then sorted by the individual ID variable.
- YULESQ
-
requests that the “Linkage Disequilibrium Measures” table be displayed and contain Yule’s
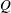 , a linkage disequilibrium measure. This option is ignored if HAPLO=NONE or NONEHWD.
, a linkage disequilibrium measure. This option is ignored if HAPLO=NONE or NONEHWD.
