The CASECONTROL Procedure
| PROC CASECONTROL Statement |
You can specify the following options in the PROC CASECONTROL statement.
- ALLELE
requests that the allele case-control test be performed. If none of the three test options (ALLELE, GENOTYPE, or TREND) are specified, then all three tests are performed by default.
- ALPHA=number
specifies that a confidence level of
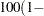 number
number 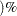 is to be used in forming confidence intervals for odds ratios. The value of number must be between 0 and 1, and is set to 0.05 by default.
is to be used in forming confidence intervals for odds ratios. The value of number must be between 0 and 1, and is set to 0.05 by default. - DATA=SAS-data-set
names the input SAS data set to be used by PROC CASECONTROL. The default is to use the most recently created data set.
- DELIMITER=’string’
indicates the string that is used to separate the two alleles that compose the genotypes contained in the variables specified in the VAR statement. This option is ignored if GENOCOL is not specified.
- GENOCOL
indicates that columns specified in the VAR statement contain genotypes instead of alleles. When this option is specified, there is one column per marker. The genotypes must consist of the two alleles separated by a delimiter . For a genotype with one missing allele, use a blank space to indicate a missing value; if both alleles are missing, either use a single missing value for the entire genotype or use the delimiter alone.
- GENOTYPE
requests that the genotype case-control test be performed. If none of the three test options (ALLELE, GENOTYPE, or TREND) are specified, then all three tests are performed by default.
- INDIVIDUAL=variable
- INDIV=variable
specifies the individual ID variable when using the TALL option. This variable can be character or numeric.
- MARKER=variable
specifies the marker ID variable when using the TALL option. This variable contains the names of the markers that are used in all output and can be character or numeric.
- NDATA=SAS-data-set
names the input SAS data set containing names, or identifiers, for the markers used in the output. There must be a NAME variable in this data set, which should contain the same number of rows as there are markers in the input data set specified in the DATA= option. When there are fewer rows than there are markers, markers without a name are named using the PREFIX= option. Likewise, if there is no NDATA= data set specified, the PREFIX= option is used. Note that this data set is ignored if the TALL option is specified in the PROC CASECONTROL statement. In that case, the marker variable names are taken from the marker ID variable specified in the MARKER= option.
- NULLSNPS=(variable list )
names the markers to be used in calculating the variance inflation factor for genomic control that is applied to the chi-square statistic(s) from the trend test. Only biallelic markers that are listed are used. Note that if GENOCOL is specified, there should be one variable for each marker listed; otherwise, there should be two variables per marker. By default, if VIF is specified in the PROC CASECONTROL statement, all biallelic markers listed in the VAR statement are used. This option must be specified if both the VIF option and the PERMS= option are used; otherwise the variance inflation factor is not applied. This option is ignored if the VIF option is not specified or if the TALL option is used.
- OR
requests that odds ratios based on allele counts for biallelic markers be included in the OUTSTAT= data set, along with (1–
 )% confidence limits for the value specified in the ALPHA=
option. Odds ratios are not reported for markers with more than two alleles.
)% confidence limits for the value specified in the ALPHA=
option. Odds ratios are not reported for markers with more than two alleles. - OUTSTAT=SAS-data-set
names the output SAS data set containing counts for the two trait values, the chi-square statistics, degrees of freedom, and
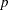 -values for the tests performed. When this option is omitted, an output data set is created by default and named according to the DATAn convention.
-values for the tests performed. When this option is omitted, an output data set is created by default and named according to the DATAn convention. - PERMS=number
indicates that Monte Carlo estimates of exact
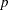 -values for the case-control tests should be calculated instead of the
-values for the case-control tests should be calculated instead of the 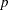 -values from the asymptotic
-values from the asymptotic 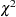 distribution. In each of the number permutation samples, the trait values are permuted among the individuals in the sample. Large values of number (10,000 or more) are usually recommended for accuracy, but long execution times can result, particularly with large data sets. When this option is omitted, no permutations are performed and
distribution. In each of the number permutation samples, the trait values are permuted among the individuals in the sample. Large values of number (10,000 or more) are usually recommended for accuracy, but long execution times can result, particularly with large data sets. When this option is omitted, no permutations are performed and 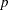 -values from the asymptotic
-values from the asymptotic 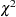 distribution are reported.
distribution are reported. - PREFIX=prefix
specifies a prefix to use in constructing names for marker variables in all output. For example, if PREFIX=VAR, the names of the variables are VAR1, VAR2, ..., VARn. Note that this option is ignored when the NDATA= option is specified, unless there are fewer names in the NDATA data set than there are markers; it is also ignored if the TALL option is specified, in which case the marker variable names are taken from the marker ID variable specified in the MARKER= option. Otherwise, if this option is omitted, PREFIX=M is the default when variables contain alleles; if GENOCOL is specified, then the names of the variables specified in the VAR statement are used as the marker names.
- SEED=number
specifies the initial seed for the random number generator used for permuting the data to calculate estimates of exact
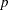 -values. This option is ignored if PERMS=
is not specified. The value for number must be an integer; the computer clock time is used if the option is omitted or an integer less than or equal to 0 is specified. For more details about seed values, see SAS Language Reference: Concepts.
-values. This option is ignored if PERMS=
is not specified. The value for number must be an integer; the computer clock time is used if the option is omitted or an integer less than or equal to 0 is specified. For more details about seed values, see SAS Language Reference: Concepts. - TALL
indicates that the input data set is of an alternative format. This tall-skinny format contains the following columns: two containing marker alleles (or one containing marker genotypes if GENOCOL is specified), one for the marker identifier, and one for the individual identifier. The MARKER= and INDIV= options must also be specified in order for this option to be in effect. Note that when this option is used, the DATA= data set must first be sorted by any BY variables, then sorted by the marker ID variable, and then sorted by the individual ID variable.
- TREND
requests that the linear trend test for allelic effects be performed. If none of the three test options (ALLELE, GENOTYPE, or TREND) are specified, then all three tests are performed by default.
- VIF
specifies that the variance inflation factor
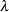 should be applied to the trend chi-square statistic for genomic control. This adjustment is applied only when the trend test is performed and to markers in the VAR statement that are biallelic.
should be applied to the trend chi-square statistic for genomic control. This adjustment is applied only when the trend test is performed and to markers in the VAR statement that are biallelic.
