The CASECONTROL Procedure
| Example |
Here are some sample SNP data on which the three case-control tests can be performed using PROC CASECONTROL:
data cc; input affected $ m1-m16; datalines; N 1 1 2 2 2 2 2 1 2 1 2 2 1 1 2 2 N 1 1 1 1 2 2 1 1 2 1 2 1 1 1 1 1 N 2 1 1 1 2 1 1 1 2 2 1 1 1 1 1 1 N 2 2 2 1 2 2 1 1 2 2 2 1 1 1 2 2 N 1 1 1 1 2 2 2 1 1 1 1 1 2 1 . . N 2 1 1 1 2 1 1 1 2 1 2 1 1 1 2 1 N 1 1 1 1 2 2 1 1 2 2 2 2 2 1 2 2 N 2 2 1 1 2 1 2 1 2 2 2 1 1 1 2 1 N 2 1 1 1 2 2 2 1 2 1 . . 1 1 2 1 N 2 1 1 1 2 1 1 1 2 2 1 1 1 1 1 1 N 2 1 2 2 . . 1 1 2 1 1 1 1 1 1 1 N 2 2 . . 2 1 1 1 2 1 2 1 1 1 2 1 N 2 1 . . 2 2 1 1 2 2 1 1 1 1 2 1 N 2 1 . . 2 2 1 1 2 1 . . 2 1 1 1 N 2 2 . . 2 2 1 1 . . 2 1 1 1 2 1 N 1 1 . . 2 2 1 1 1 1 2 1 1 1 2 1 N 1 1 . . 2 2 1 1 1 1 . . 1 1 2 1 N 2 1 . . 2 2 1 1 1 1 . . 2 1 2 1 A 2 1 2 1 2 1 1 1 1 1 2 1 . . 2 1 A 2 1 2 1 2 2 1 1 2 1 1 1 . . 1 1 A 2 2 2 1 2 2 1 1 2 2 . . . . 2 1 A 2 1 2 2 2 1 1 1 2 1 2 1 . . 2 2 A . . 2 2 2 1 . . 1 1 2 2 . . 2 1 A 1 1 1 1 2 1 1 1 2 1 1 1 . . 2 2 A 2 1 1 1 2 2 1 1 1 1 2 1 . . 2 1 A 2 1 2 2 2 2 1 1 2 2 . . . . 2 2 A 2 1 1 1 2 2 1 1 2 1 2 1 . . 1 1 A 2 1 2 2 2 1 1 1 2 1 2 1 . . 2 2 A 1 1 1 1 2 2 1 1 2 1 2 1 . . 2 2 A 2 1 2 1 2 1 1 1 2 1 2 2 . . 2 1 A 2 2 2 2 1 1 1 1 2 1 2 1 . . 2 2 A 1 1 1 1 2 1 . . 2 1 2 2 . . 2 2 A 1 1 2 1 2 1 1 1 2 1 2 1 . . 2 2 A 2 2 1 1 2 2 1 1 2 1 1 1 . . 2 1 ;
The following SAS code can be used to perform the analysis:
proc casecontrol data=cc prefix=Marker; var m1-m16; trait affected; run;
proc print heading=h; format probgenotype proballele probtrend pvalue5.4; format chisqgenotype chisqallele chisqtrend 5.3; run;
All three case-control tests are performed by default. The output data set created by default appears in Figure 5.1.
| Obs | Locus | NumTraitA | NumTraitN | ChiSqGenotype | ChiSqAllele | ChiSqTrend | dfGenotype | dfAllele | dfTrend | ProbGenotype | ProbAllele | ProbTrend |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Marker1 | 15 | 18 | 0.272 | 0.033 | 0.032 | 2 | 1 | 1 | 0.873 | 0.857 | 0.858 |
| 2 | Marker2 | 16 | 11 | 3.430 | 3.260 | 2.140 | 2 | 1 | 1 | 0.180 | 0.071 | 0.144 |
| 3 | Marker3 | 16 | 17 | 2.981 | 2.569 | 2.925 | 2 | 1 | 1 | 0.225 | 0.109 | 0.087 |
| 4 | Marker4 | 14 | 18 | 3.556 | 3.319 | 3.556 | 1 | 1 | 1 | 0.059 | 0.069 | 0.059 |
| 5 | Marker5 | 16 | 17 | 3.004 | 0.535 | 0.590 | 2 | 1 | 1 | 0.223 | 0.464 | 0.443 |
| 6 | Marker6 | 14 | 14 | 0.767 | 0.650 | 0.710 | 2 | 1 | 1 | 0.682 | 0.420 | 0.399 |
| 7 | Marker7 | 0 | 18 | 0.000 | 0.000 | 0.000 | 0 | 0 | 0 | . | . | . |
| 8 | Marker8 | 16 | 17 | 4.132 | 4.061 | 3.769 | 2 | 1 | 1 | 0.127 | 0.044 | 0.052 |
Figure 5.1 displays the statistics for the three tests. The genotype case-control statistic has more degrees of freedom than the other two because it is testing for both dominance genotypic effects and additive allelic effects, while the other statistics are testing for the significant additive effects alone. Using the standard significance level of 0.05, none of the 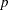 -values, shown in the last three columns, would be considered significant since they are all above this significance level. Thus, you would conclude that none of the markers show a significant association with the binary trait. The
-values, shown in the last three columns, would be considered significant since they are all above this significance level. Thus, you would conclude that none of the markers show a significant association with the binary trait. The 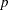 -values for Marker7 are missing because the genotypes of all the affected individuals are missing at that marker.
-values for Marker7 are missing because the genotypes of all the affected individuals are missing at that marker.
