The BTL Procedure (Experimental)
Overview: BTL Procedure
The BTL procedure analyzes marker and trait data in order to find and characterize binary trait loci (BTL). Mixed model analysis of variance is used to find a locus or loci associated with a trait, and a maximum likelihood model is used to estimate the recombination and penetrance parameters for a given set of BTL.
The data consist of marker genotype and a binary trait for a set of individuals. Marker genotypes can be of single nucleotide polymorphisms (SNPs), microsatellite data, or any other kind of marker as long as it is a heritable unit that obeys the laws of transmission genetics. However, only biallelic markers can be used to estimate BTL parameters; multiallelic markers can be used to find BTL but not to estimate parameters.
The output of PROC BTL is a "Model Statistics" table that contains a sorted list of the BTL models evaluated by a user-specified model selection criterion (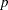 -value by default). Additionally, the maximum likelihood parameter estimates for a selected BTL model are written to a "Parameter Estimates" table if the PARMEST statement is used. The penetrance parameters can be calculated for a specified set of recombination parameters, or alternatively, a grid search can be performed over a specified range of possible recombination parameters. Finally, a
-value by default). Additionally, the maximum likelihood parameter estimates for a selected BTL model are written to a "Parameter Estimates" table if the PARMEST statement is used. The penetrance parameters can be calculated for a specified set of recombination parameters, or alternatively, a grid search can be performed over a specified range of possible recombination parameters. Finally, a 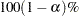 confidence interval can be computed for the parameter estimates of a given model.
confidence interval can be computed for the parameter estimates of a given model.
Note: This procedure is experimental.
