The DISCRIM Procedure
Example 35.1 Univariate Density Estimates and Posterior Probabilities
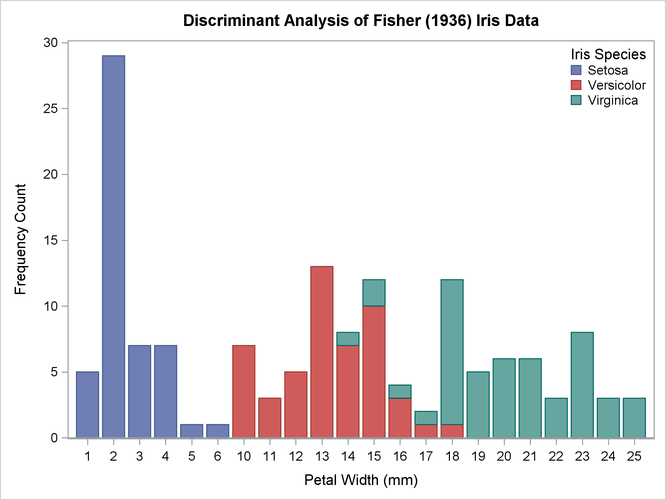
In this example, several discriminant analyses are run with a single quantitative variable, petal width, so that density estimates and posterior probabilities can be plotted easily. The example produces Output 35.1.1 through Output 35.1.5. ODS Graphics is used to display the sample distribution of petal width in the three species. For general information about ODS Graphics, see Chapter 21: Statistical Graphics Using ODS. Note the overlap between the species I. versicolor and I. virginica that the bar chart shows. The following statements produce Output 35.1.1:
title 'Discriminant Analysis of Fisher (1936) Iris Data'; proc freq data=sashelp.iris noprint; tables petalwidth * species / out=freqout; run; proc sgplot data=freqout; vbar petalwidth / response=count group=species; keylegend / location=inside position=ne noborder across=1; run;
Output 35.1.1: Sample Distribution of Petal Width in Three Species
In order to plot the density estimates and posterior probabilities, a data set called plotdata is created containing equally spaced values from –5 to 30, covering the range of petal width with a little to spare on each
end. The plotdata data set is used with the TESTDATA= option in PROC DISCRIM. The following statements make the data set:
data plotdata;
do PetalWidth=-5 to 30 by 0.5;
output;
end;
run;
The same plots are produced after each discriminant analysis, so macros are used to reduce the amount of typing required.
The macros use two data sets. The data set plotd, containing density estimates, is created by the TESTOUTD= option in PROC DISCRIM. The data set plotp, containing posterior probabilities, is created by the TESTOUT= option. For each data set, the macros remove uninteresting
values (near zero) and create an overlay plot showing all three species in a single plot.
The following statements create the macros:
%macro plotden;
title3 'Plot of Estimated Densities';
data plotd2;
set plotd;
if setosa < .002 then setosa = .;
if versicolor < .002 then versicolor = .;
if virginica < .002 then virginica = .;
g = 'Setosa '; Density = setosa; output;
g = 'Versicolor'; Density = versicolor; output;
g = 'Virginica '; Density = virginica; output;
label PetalWidth='Petal Width in mm.';
run;
proc sgplot data=plotd2;
series y=Density x=PetalWidth / group=g;
discretelegend;
run;
%mend;
%macro plotprob;
title3 'Plot of Posterior Probabilities';
data plotp2;
set plotp;
if setosa < .01 then setosa = .;
if versicolor < .01 then versicolor = .;
if virginica < .01 then virginica = .;
g = 'Setosa '; Probability = setosa; output;
g = 'Versicolor'; Probability = versicolor; output;
g = 'Virginica '; Probability = virginica; output;
label PetalWidth='Petal Width in mm.';
run;
proc sgplot data=plotp2;
series y=Probability x=PetalWidth / group=g;
discretelegend;
run;
%mend;
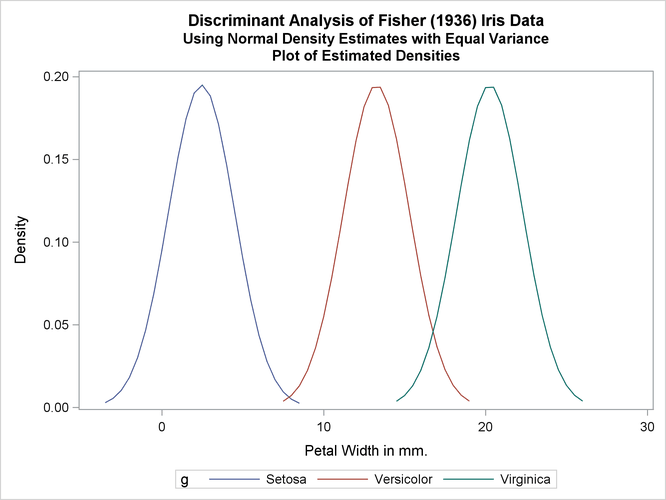
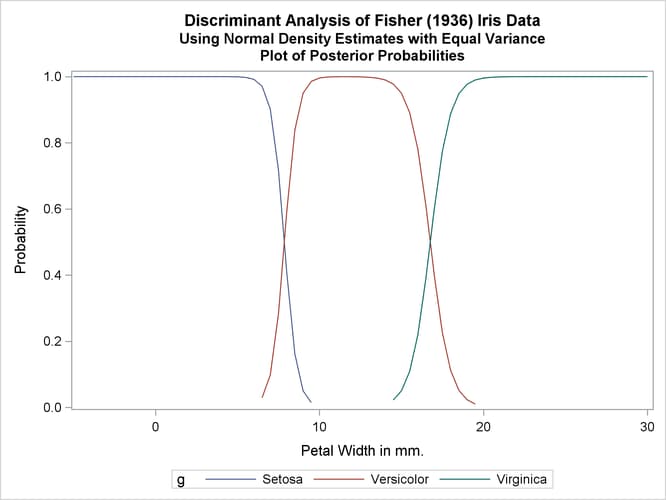
The first analysis uses normal-theory methods (METHOD=NORMAL) assuming equal variances (POOL=YES) in the three classes. The NOCLASSIFY option suppresses the resubstitution classification results of the input data set observations. The CROSSLISTERR option lists the observations that are misclassified under cross validation and displays cross validation error-rate estimates. The following statements produce Output 35.1.2:
title2 'Using Normal Density Estimates with Equal Variance';
proc discrim data=sashelp.iris method=normal pool=yes
testdata=plotdata testout=plotp testoutd=plotd
short noclassify crosslisterr;
class Species;
var PetalWidth;
run;
%plotden;
%plotprob;
Output 35.1.2: Normal Density Estimates with Equal Variance
| Discriminant Analysis of Fisher (1936) Iris Data |
| Using Normal Density Estimates with Equal Variance |
| Posterior Probability of Membership in Species | ||||||
|---|---|---|---|---|---|---|
| Obs | From Species | Classified into Species |
Setosa | Versicolor | Virginica | |
| 53 | Versicolor | Virginica | * | 0.0000 | 0.0952 | 0.9048 |
| 100 | Versicolor | Virginica | * | 0.0000 | 0.3828 | 0.6172 |
| 103 | Virginica | Versicolor | * | 0.0000 | 0.9610 | 0.0390 |
| 124 | Virginica | Versicolor | * | 0.0000 | 0.9940 | 0.0060 |
| 130 | Virginica | Versicolor | * | 0.0000 | 0.8009 | 0.1991 |
| 136 | Virginica | Versicolor | * | 0.0000 | 0.9610 | 0.0390 |
| * Misclassified observation |
| Discriminant Analysis of Fisher (1936) Iris Data |
| Using Normal Density Estimates with Equal Variance |
| Number of Observations and Percent Classified into Species |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| From Species | Setosa | Versicolor | Virginica | Total | ||||||||
| Setosa |
|
|
|
|
||||||||
| Versicolor |
|
|
|
|
||||||||
| Virginica |
|
|
|
|
||||||||
| Total |
|
|
|
|
||||||||
| Priors |
|
|
|
|
||||||||
| Discriminant Analysis of Fisher (1936) Iris Data |
| Using Normal Density Estimates with Equal Variance |
| Observation Profile for Test Data | |
|---|---|
| Number of Observations Read | 71 |
| Number of Observations Used | 71 |
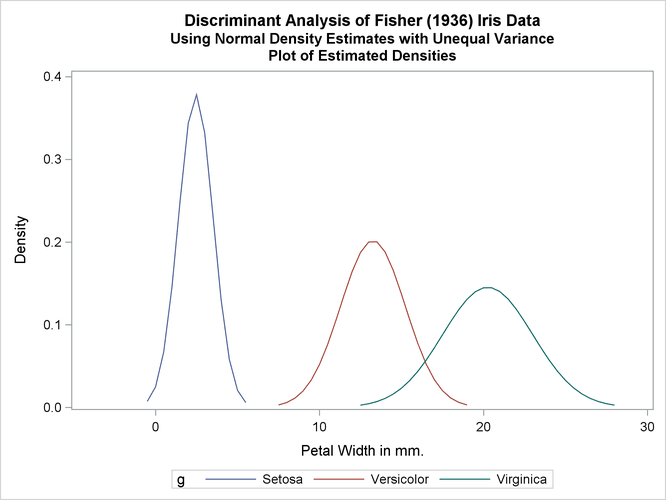
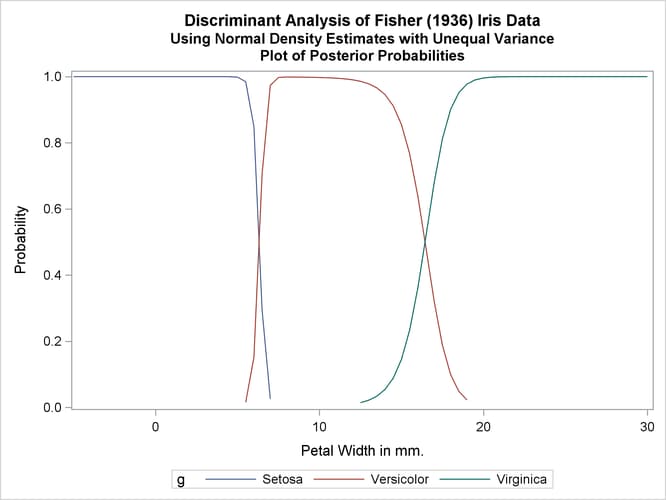
The next analysis uses normal-theory methods assuming unequal variances (POOL=NO) in the three classes. The following statements produce Output 35.1.3:
title2 'Using Normal Density Estimates with Unequal Variance';
proc discrim data=sashelp.iris method=normal pool=no
testdata=plotdata testout=plotp testoutd=plotd
short noclassify crosslisterr;
class Species;
var PetalWidth;
run;
%plotden;
%plotprob;
Output 35.1.3: Normal Density Estimates with Unequal Variance
| Discriminant Analysis of Fisher (1936) Iris Data |
| Using Normal Density Estimates with Unequal Variance |
| Posterior Probability of Membership in Species | ||||||
|---|---|---|---|---|---|---|
| Obs | From Species | Classified into Species |
Setosa | Versicolor | Virginica | |
| 10 | Setosa | Versicolor | * | 0.4923 | 0.5073 | 0.0004 |
| 53 | Versicolor | Virginica | * | 0.0000 | 0.0686 | 0.9314 |
| 100 | Versicolor | Virginica | * | 0.0000 | 0.2871 | 0.7129 |
| 103 | Virginica | Versicolor | * | 0.0000 | 0.8740 | 0.1260 |
| 124 | Virginica | Versicolor | * | 0.0000 | 0.9602 | 0.0398 |
| 130 | Virginica | Versicolor | * | 0.0000 | 0.6558 | 0.3442 |
| 136 | Virginica | Versicolor | * | 0.0000 | 0.8740 | 0.1260 |
| * Misclassified observation |
| Discriminant Analysis of Fisher (1936) Iris Data |
| Using Normal Density Estimates with Unequal Variance |
| Number of Observations and Percent Classified into Species |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| From Species | Setosa | Versicolor | Virginica | Total | ||||||||
| Setosa |
|
|
|
|
||||||||
| Versicolor |
|
|
|
|
||||||||
| Virginica |
|
|
|
|
||||||||
| Total |
|
|
|
|
||||||||
| Priors |
|
|
|
|
||||||||
| Discriminant Analysis of Fisher (1936) Iris Data |
| Using Normal Density Estimates with Unequal Variance |
| Observation Profile for Test Data | |
|---|---|
| Number of Observations Read | 71 |
| Number of Observations Used | 71 |
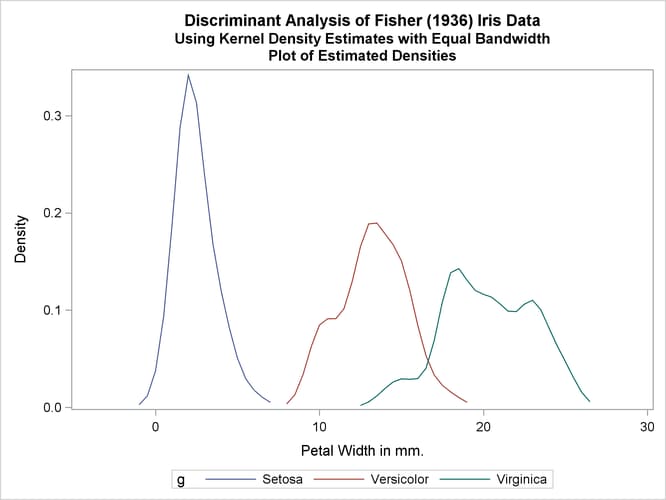
Two more analyses are run with nonparametric methods (METHOD=NPAR), specifically kernel density estimates with normal kernels (KERNEL=NORMAL). The first of these uses equal bandwidths (smoothing parameters) (POOL=YES) in each class. The use of equal bandwidths does not constrain the density estimates to be of equal variance. The value of the radius parameter that, assuming normality, minimizes an approximate mean integrated square error is 0.48 (see the section Nonparametric Methods). Choosing r = 0.4 gives a more detailed look at the irregularities in the data. The following statements produce Output 35.1.4:
title2 'Using Kernel Density Estimates with Equal Bandwidth';
proc discrim data=sashelp.iris method=npar kernel=normal
r=.4 pool=yes
testdata=plotdata testout=plotp
testoutd=plotd
short noclassify crosslisterr;
class Species;
var PetalWidth;
run;
%plotden;
%plotprob;
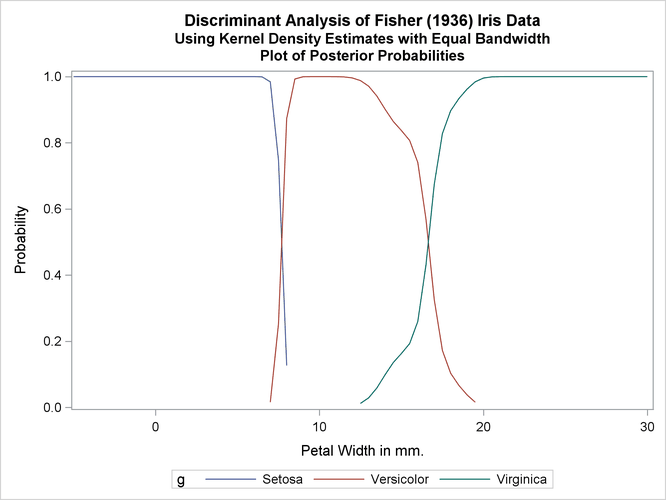
Output 35.1.4: Kernel Density Estimates with Equal Bandwidth
| Discriminant Analysis of Fisher (1936) Iris Data |
| Using Kernel Density Estimates with Equal Bandwidth |
| Posterior Probability of Membership in Species | ||||||
|---|---|---|---|---|---|---|
| Obs | From Species | Classified into Species |
Setosa | Versicolor | Virginica | |
| 53 | Versicolor | Virginica | * | 0.0000 | 0.0438 | 0.9562 |
| 100 | Versicolor | Virginica | * | 0.0000 | 0.2586 | 0.7414 |
| 103 | Virginica | Versicolor | * | 0.0000 | 0.8827 | 0.1173 |
| 124 | Virginica | Versicolor | * | 0.0000 | 0.9472 | 0.0528 |
| 130 | Virginica | Versicolor | * | 0.0000 | 0.8061 | 0.1939 |
| 136 | Virginica | Versicolor | * | 0.0000 | 0.8827 | 0.1173 |
| * Misclassified observation |
| Discriminant Analysis of Fisher (1936) Iris Data |
| Using Kernel Density Estimates with Equal Bandwidth |
| Number of Observations and Percent Classified into Species |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| From Species | Setosa | Versicolor | Virginica | Total | ||||||||
| Setosa |
|
|
|
|
||||||||
| Versicolor |
|
|
|
|
||||||||
| Virginica |
|
|
|
|
||||||||
| Total |
|
|
|
|
||||||||
| Priors |
|
|
|
|
||||||||
| Discriminant Analysis of Fisher (1936) Iris Data |
| Using Kernel Density Estimates with Equal Bandwidth |
| Observation Profile for Test Data | |
|---|---|
| Number of Observations Read | 71 |
| Number of Observations Used | 71 |
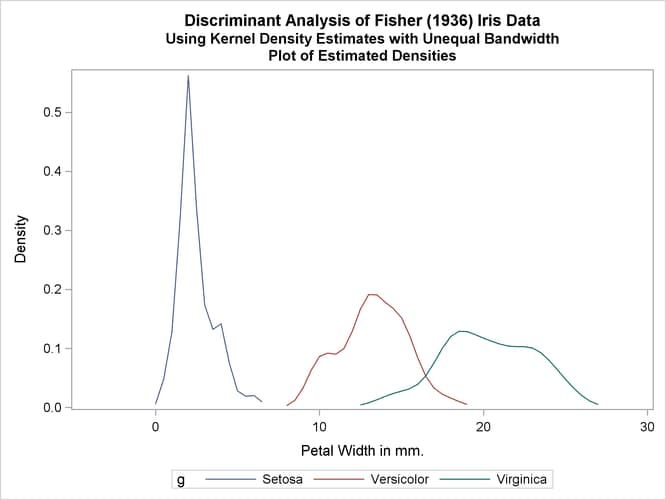
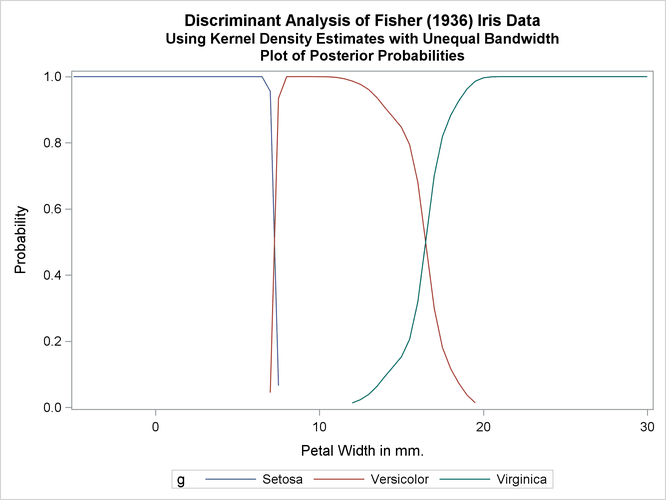
Another nonparametric analysis is run with unequal bandwidths (POOL=NO). The following statements produce Output 35.1.5:
title2 'Using Kernel Density Estimates with Unequal Bandwidth';
proc discrim data=sashelp.iris method=npar kernel=normal
r=.4 pool=no
testdata=plotdata testout=plotp
testoutd=plotd
short noclassify crosslisterr;
class Species;
var PetalWidth;
run;
%plotden;
%plotprob;
Output 35.1.5: Kernel Density Estimates with Unequal Bandwidth
| Discriminant Analysis of Fisher (1936) Iris Data |
| Using Kernel Density Estimates with Unequal Bandwidth |
| Posterior Probability of Membership in Species | ||||||
|---|---|---|---|---|---|---|
| Obs | From Species | Classified into Species |
Setosa | Versicolor | Virginica | |
| 53 | Versicolor | Virginica | * | 0.0000 | 0.0475 | 0.9525 |
| 100 | Versicolor | Virginica | * | 0.0000 | 0.2310 | 0.7690 |
| 103 | Virginica | Versicolor | * | 0.0000 | 0.8805 | 0.1195 |
| 124 | Virginica | Versicolor | * | 0.0000 | 0.9394 | 0.0606 |
| 130 | Virginica | Versicolor | * | 0.0000 | 0.7193 | 0.2807 |
| 136 | Virginica | Versicolor | * | 0.0000 | 0.8805 | 0.1195 |
| * Misclassified observation |
| Discriminant Analysis of Fisher (1936) Iris Data |
| Using Kernel Density Estimates with Unequal Bandwidth |
| Number of Observations and Percent Classified into Species |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| From Species | Setosa | Versicolor | Virginica | Total | ||||||||
| Setosa |
|
|
|
|
||||||||
| Versicolor |
|
|
|
|
||||||||
| Virginica |
|
|
|
|
||||||||
| Total |
|
|
|
|
||||||||
| Priors |
|
|
|
|
||||||||
| Discriminant Analysis of Fisher (1936) Iris Data |
| Using Kernel Density Estimates with Unequal Bandwidth |
| Observation Profile for Test Data | |
|---|---|
| Number of Observations Read | 71 |
| Number of Observations Used | 71 |