The ICLIFETEST Procedure
Suppose the event times for a total of n subjects, ![]() ,
, ![]() , …,
, …, ![]() , are independent random variables with an underlying cumulative distribution function
, are independent random variables with an underlying cumulative distribution function ![]() . Denote the corresponding survival function as
. Denote the corresponding survival function as ![]() . Interval-censoring occurs when some or all
. Interval-censoring occurs when some or all ![]() ’s cannot be observed directly but are known to be within the interval
’s cannot be observed directly but are known to be within the interval ![]() .
.
The observed intervals might or might not overlap. It they do not overlap, then you can usually use conventional methods for right-censored data, with minor modifications. On the other hand, if some intervals overlap, you need special algorithms to compute an unbiased estimate of the underlying survival function.
To characterize the nonparametric estimate of the survival function, Peto (1973) and Turnbull (1976) show that the estimate can jump only at the right endpoint of a set of nonoverlapping intervals (also known as Turnbull
intervals), ![]() . A simple algorithm for finding these intervals is to order all the boundary values
. A simple algorithm for finding these intervals is to order all the boundary values ![]() with labels of L and R attached and then pick up the intervals that have L as the left boundary and R as the right boundary. For example, suppose that the data set contains only three intervals,
with labels of L and R attached and then pick up the intervals that have L as the left boundary and R as the right boundary. For example, suppose that the data set contains only three intervals, ![]() ,
, ![]() , and
, and ![]() . The ordered values are
. The ordered values are ![]() . Then the Turnbull intervals are
. Then the Turnbull intervals are ![]() and
and ![]() .
.
For the exact observation ![]() , Ng (2002) suggests that it be represented by the interval
, Ng (2002) suggests that it be represented by the interval ![]() for a positive small value
for a positive small value ![]() . If
. If ![]() for an observation
for an observation ![]() (
(![]() ), then the observation is represented by
), then the observation is represented by ![]() .
.
Define ![]() ,
, ![]() . Given the data, the survival function,
. Given the data, the survival function, ![]() , can be determined only up to equivalence classes t, which are complements of the Turnbull intervals.
, can be determined only up to equivalence classes t, which are complements of the Turnbull intervals. ![]() is undefined if t is within some
is undefined if t is within some ![]() . The likelihood function for
. The likelihood function for ![]() is then
is then
where ![]() is 1 if
is 1 if ![]() is contained in
is contained in ![]() and 0 otherwise.
and 0 otherwise.
Denote the maximum likelihood estimate for ![]() as
as ![]() . The survival function can then be estimated as
. The survival function can then be estimated as
Peto (1973) suggests maximizing this likelihood function by using a Newton-Raphson algorithm subject to the constraint ![]() . This approach has been implemented in the
. This approach has been implemented in the ![]() ICE macro. Although feasible, the optimization becomes less stable as the dimension of
ICE macro. Although feasible, the optimization becomes less stable as the dimension of ![]() increases.
increases.
Treating interval-censored data as missing data, Turnbull (1976) derives a self-consistent equation for estimating the ![]() ’s:
’s:
where ![]() is the expected probability that the event
is the expected probability that the event ![]() occurs within
occurs within ![]() for the ith subject, given the observed data.
for the ith subject, given the observed data.
The algorithm is an expectation-maximization (EM) algorithm in the sense that it iteratively updates ![]() and
and ![]() . Convergence is declared if, for a chosen number
. Convergence is declared if, for a chosen number ![]() ,
,
where ![]() denotes the updated value for
denotes the updated value for ![]() after the lth iteration.
after the lth iteration.
An alternative criterion is to declare convergence when increments of the likelihood are small:
There is no guarantee that the converged values constitute a maximum likelihood estimate (MLE). Gentleman and Geyer (1994) introduced the Kuhn-Tucker conditions based on constrained programming as a check of whether the algorithm converges to
a legitimate MLE. These conditions state that a sufficient and necessary condition for the estimate to be a MLE is that the
Lagrange multipliers ![]() are nonnegative for all the
are nonnegative for all the ![]() ’s that are estimated to be zero, where
’s that are estimated to be zero, where ![]() is the derivative of the log-likelihood function with respect to
is the derivative of the log-likelihood function with respect to ![]() :
:
You can use Turnbull’s method by specifying METHOD=TURNBULL in the ICLIFETEST statement. The Lagrange multipliers are displayed in the "Nonparametric Survival Estimates" table.
Groeneboom and Wellner (1992) propose using the iterative convex minorant (ICM) algorithm to estimate the underlying survival function as an alternative
to Turnbull’s method. Define ![]() ,
, ![]() as the cumulative probability at the right boundary of the jth Turnbull interval:
as the cumulative probability at the right boundary of the jth Turnbull interval: ![]() . It follows that
. It follows that ![]() . Denote
. Denote ![]() and
and ![]() . You can rewrite the likelihood function as
. You can rewrite the likelihood function as
Maximizing the likelihood with respect to the ![]() ’s is equivalent to maximizing it with respect to the
’s is equivalent to maximizing it with respect to the ![]() ’s. Because the
’s. Because the ![]() ’s are naturally ordered, the optimization is subject to the following constraint:
’s are naturally ordered, the optimization is subject to the following constraint:
Denote the log-likelihood function as ![]() . Suppose its maximum occurs at
. Suppose its maximum occurs at ![]() . Mathematically, it can be proved that
. Mathematically, it can be proved that ![]() equals the maximizer of the following quadratic function:
equals the maximizer of the following quadratic function:
where ![]() ,
, ![]() denotes the derivatives of
denotes the derivatives of ![]() with respect to
with respect to ![]() , and
, and ![]() is a positive definite matrix of size
is a positive definite matrix of size ![]() (Groeneboom and Wellner, 1992).
(Groeneboom and Wellner, 1992).
An iterative algorithm is needed to determine ![]() . For the lth iteration, the algorithm updates the quantity
. For the lth iteration, the algorithm updates the quantity
where ![]() is the parameter estimate from the previous iteration and
is the parameter estimate from the previous iteration and ![]() is a positive definite diagonal matrix that depends on
is a positive definite diagonal matrix that depends on ![]() .
.
A convenient choice for ![]() is the negative of the second-order derivative of the log-likelihood function
is the negative of the second-order derivative of the log-likelihood function ![]() :
:
Given ![]() and
and ![]() , the parameter estimate for the lth iteration
, the parameter estimate for the lth iteration ![]() maximizes the quadratic function
maximizes the quadratic function ![]() .
.
Define the cumulative sum diagram ![]() as a set of m points in the plane, where
as a set of m points in the plane, where ![]() and
and
Technically, ![]() equals the left derivative of the convex minorant, or in other words, the largest convex function below the diagram
equals the left derivative of the convex minorant, or in other words, the largest convex function below the diagram ![]() . This optimization problem can be solved by the pool-adjacent-violators algorithm (Groeneboom and Wellner, 1992).
. This optimization problem can be solved by the pool-adjacent-violators algorithm (Groeneboom and Wellner, 1992).
Occasionally, the ICM step might not increase the likelihood. Jongbloed (1998) suggests conducting a line search to ensure that positive increments are always achieved. Alternatively, you can switch to the EM step, exploiting the fact that the EM iteration never decreases the likelihood, and then resume iterations of the ICM algorithm after the EM step. As with Turnbull’s method, convergence can be determined based on the closeness of two consecutive sets of parameter values or likelihood values. You can use the ICM algorithm by specifying METHOD=ICM in the PROC ICLIFETEST statement.
As its name suggests, the EMICM algorithm combines the self-consistent EM algorithm and the ICM algorithm by alternating the two different steps in its iterations. Wellner and Zhan (1997) show that the converged values of the EMICM algorithm always constitute an MLE if it exists and is unique. The ICLIFETEST procedure uses the EMICM algorithm as the default.
Peto (1973) and Turnbull (1976) suggest estimating the variances of the survival estimates by inverting the Hessian matrix, which is obtained by twice differentiating
the log-likelihood function. This method can become less stable when the number of ![]() ’s increase as n increases. Simulations have shown that the confidence limits based on variances estimated with this method tend to have conservative
coverage probabilities that are greater than the nominal level (Goodall, Dunn, and Babiker, 2004).
’s increase as n increases. Simulations have shown that the confidence limits based on variances estimated with this method tend to have conservative
coverage probabilities that are greater than the nominal level (Goodall, Dunn, and Babiker, 2004).
Sun (2001) proposes using two resampling techniques, simple bootstrap and multiple imputation, to estimate the variance of the survival estimator. The undefined regions that the Turnbull intervals represent create a special challenge using the bootstrap method. Because each bootstrap sample could have a different set of Turnbull intervals, some time points to evaluate the variances based on the original Turnbull intervals might be located within the intervals in a bootstrap sample, with the result that their survival probabilities become unknown. A simple ad hoc solution is to shrink the Turnbull interval to its right boundary and modify the survival estimates into a right continuous function:
Let M denote the number of resampling data sets. Let ![]() denote the n independent samples from the original data with replacement,
denote the n independent samples from the original data with replacement, ![]() . Let
. Let ![]() be the modified estimate of the survival function computed from the kth resampling data set. Then you can estimate the variance of
be the modified estimate of the survival function computed from the kth resampling data set. Then you can estimate the variance of ![]() by the sample variance as
by the sample variance as
where
The method of multiple imputations exploits the fact that interval-censored data reduce to right-censored data when all interval
observations of finite length shrink to single points. Suppose that each finite interval has been converted to one of the
![]() values it contains. For this right-censored data set, you can estimate the variance of the survival estimates via the well-known
Greenwood formula (1958) as
values it contains. For this right-censored data set, you can estimate the variance of the survival estimates via the well-known
Greenwood formula (1958) as
where ![]() is the number of events at time
is the number of events at time ![]() and
and ![]() is the number of subjects at risk just prior to
is the number of subjects at risk just prior to ![]() , and
, and ![]() is the Kaplan-Meier estimator of the survival function,
is the Kaplan-Meier estimator of the survival function,
Essentially, multiple imputation is used to account for the uncertainty of ranking overlapping intervals. The kth imputed data set is obtained by substituting every interval-censored observation of finite length with an exact event time randomly drawn from the conditional survival function:
Because ![]() only jumps at the
only jumps at the ![]() , this is a discrete function.
, this is a discrete function.
Denote the Kaplan-Meier estimate of each imputed data set as ![]() . The variance of
. The variance of ![]() is estimated by
is estimated by
where
and
and
Note that the first term in the formula for ![]() mimics the Greenwood formula but uses expected numbers of deaths and subjects. The second term is the sample variance of
the Kaplan-Meier estimates of imputed data sets, which accounts for between-imputation contributions.
mimics the Greenwood formula but uses expected numbers of deaths and subjects. The second term is the sample variance of
the Kaplan-Meier estimates of imputed data sets, which accounts for between-imputation contributions.
Pointwise confidence limits can be computed for the survival function given the estimated standard errors. Let ![]() be specified by the ALPHA= option. Let
be specified by the ALPHA= option. Let ![]() be the critical value for the standard normal distribution. That is,
be the critical value for the standard normal distribution. That is, ![]() , where
, where ![]() is the cumulative distribution function of the standard normal random variable.
is the cumulative distribution function of the standard normal random variable.
Constructing the confidence limits for the survival function ![]() as
as ![]() might result in an estimate that exceeds the range [0,1] at extreme values of t. This problem can be avoided by applying a transformation to
might result in an estimate that exceeds the range [0,1] at extreme values of t. This problem can be avoided by applying a transformation to ![]() so that the range is unrestricted. In addition, certain transformed confidence intervals for
so that the range is unrestricted. In addition, certain transformed confidence intervals for ![]() perform better than the usual linear confidence intervals (Borgan and Liestøl, 1990). You can use the CONFTYPE= option to set one of the following transformations: the log-log function (Kalbfleisch and Prentice,
1980), the arcsine–square root function (Nair, 1984), the logit function (Meeker and Escobar, 1998), the log function, and the linear function.
perform better than the usual linear confidence intervals (Borgan and Liestøl, 1990). You can use the CONFTYPE= option to set one of the following transformations: the log-log function (Kalbfleisch and Prentice,
1980), the arcsine–square root function (Nair, 1984), the logit function (Meeker and Escobar, 1998), the log function, and the linear function.
Let g denote the transformation that is being applied to the survival function ![]() . Using the delta method, you estimate the standard error of
. Using the delta method, you estimate the standard error of ![]() by
by
where g’ is the first derivative of the function g. The 100(1 – ![]() )% confidence interval for
)% confidence interval for ![]() is given by
is given by
where ![]() is the inverse function of g. The choices for the transformation g are as follows:
is the inverse function of g. The choices for the transformation g are as follows:
-
arcsine–square root transformation: The estimated variance of
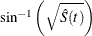 is
is ![$ \hat{\tau }^2(t) = \frac{\hat{\sigma }^2[\hat{S}(t)]}{4\hat{S}(t)[1- \hat{S}(t)] }. $](images/statug_iclifetest0135.png) The 100(1 –
The 100(1 –  )% confidence interval for
)% confidence interval for 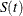 is given by
is given by
![\[ \sin ^2\left\{ \max \left[0,\sin ^{-1}(\sqrt {\hat{S}(t)}) - z_{\frac{\alpha }{2}} \hat{\tau }(t)\right]\right\} \le S(t) \le \sin ^2\left\{ \min \left[\frac{\pi }{2},\sin ^{-1}(\sqrt {\hat{S}(t)}) + z_{\frac{\alpha }{2}} \hat{\tau }(t)\right]\right\} \]](images/statug_iclifetest0136.png)
-
linear transformation: This is the same as the identity transformation. The 100(1 –
 )% confidence interval for
)% confidence interval for 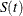 is given by
is given by
![\[ \hat{S}(t) - z_{\frac{\alpha }{2}}\hat{\sigma }\left[\hat{S}(t)\right] \le S(t) \le \hat{S}(t) + z_{\frac{\alpha }{2}}\hat{\sigma }\left[\hat{S}(t)\right] \]](images/statug_iclifetest0137.png)
-
log transformation: The estimated variance of
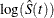 is
is 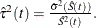 The 100(1 –
The 100(1 –  )% confidence interval for
)% confidence interval for 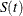 is given by
is given by
![\[ \hat{S}(t)\exp \left(-z_{\frac{\alpha }{2}}\hat{\tau }(t)\right) \le S(t) \le \hat{S}(t)\exp \left(z_{\frac{\alpha }{2}}\hat{\tau }(t)\right) \]](images/statug_iclifetest0140.png)
-
log-log transformation: The estimated variance of
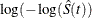 is
is ![$ \hat{\tau }^2(t) = \frac{\hat{\sigma }^2[\hat{S}(t)]}{ [\hat{S}(t)\log (\hat{S}(t))]^2 }. $](images/statug_iclifetest0142.png) The 100(1 –
The 100(1 –  )% confidence interval for
)% confidence interval for 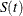 is given by
is given by
![\[ \left[\hat{S}(t)\right]^{\exp \left( z_{\frac{\alpha }{2}} \hat{\tau }(t)\right)} \le S(t) \le \left[\hat{S}(t)\right]^{\exp \left(-z_{\frac{\alpha }{2}} \hat{\tau }(t)\right)} \]](images/statug_iclifetest0143.png)
-
logit transformation: The estimated variance of
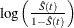 is
is
![\[ \hat{\tau }^2(t) = \frac{\hat{\sigma }^2(\hat{S}(t))}{\hat{S}^2(t)[1- \hat{S}(t)]^2} \]](images/statug_iclifetest0145.png)
The 100(1 –
 )% confidence limits for
)% confidence limits for 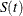 are given by
are given by
![\[ \frac{\hat{S}(t)}{\hat{S}(t) + \left[1 -\hat{S}(t) \right] \exp \left(z_{\frac{\alpha }{2}}\hat{\tau }(t)\right)} \le S(t) \le \frac{\hat{S}(t)}{\hat{S}(t) + \left[1 -\hat{S}(t) \right] \exp \left(-z_{\frac{\alpha }{2}}\hat{\tau }(t)\right)} \]](images/statug_iclifetest0146.png)
The first quartile (25th percentile) of the survival time is the time beyond which 75% of the subjects in the population under
study are expected to survive. For interval-censored data, it is problematic to define point estimators of the quartiles based
on the survival estimate ![]() because of its undefined regions of Turnbull intervals. To overcome this problem, you need to impute survival probabilities
within the Turnbull intervals. The previously defined estimator
because of its undefined regions of Turnbull intervals. To overcome this problem, you need to impute survival probabilities
within the Turnbull intervals. The previously defined estimator ![]() achieves this by placing all the estimated probabilities at the right boundary of the interval. The first quartile is estimated
by
achieves this by placing all the estimated probabilities at the right boundary of the interval. The first quartile is estimated
by
If ![]() is exactly equal to 0.75 from
is exactly equal to 0.75 from ![]() to
to ![]() , the first quartile is taken to be
, the first quartile is taken to be ![]() . If
. If ![]() is greater than 0.75 for all values of t, the first quartile cannot be estimated and is represented by a missing value in the printed output.
is greater than 0.75 for all values of t, the first quartile cannot be estimated and is represented by a missing value in the printed output.
The general formula for estimating the 100p percentile point is
The second quartile (the median) and the third quartile of survival times correspond to p = 0.5 and p = 0.75, respectively.
Brookmeyer and Crowley (1982) constructed the confidence interval for the median survival time based on the confidence interval for the survival function
![]() . The methodology is generalized to construct the confidence interval for the 100p percentile based on a g-transformed confidence interval for
. The methodology is generalized to construct the confidence interval for the 100p percentile based on a g-transformed confidence interval for ![]() (Klein and Moeschberger, 1997). You can use the CONFTYPE= option to specify the g-transformation. The
(Klein and Moeschberger, 1997). You can use the CONFTYPE= option to specify the g-transformation. The ![]() % confidence interval for the first quantile survival time is the set of all points t that satisfy
% confidence interval for the first quantile survival time is the set of all points t that satisfy
where ![]() is the first derivative of
is the first derivative of ![]() and
and ![]() is the
is the ![]() percentile of the standard normal distribution.
percentile of the standard normal distribution.
After you obtain the survival estimate ![]() , you can construct a discrete estimator for the cumulative hazard function. First, you compute the jumps of the discrete
function as
, you can construct a discrete estimator for the cumulative hazard function. First, you compute the jumps of the discrete
function as
where the ![]() ’s have been defined previously for calculating the Lagrange multiplier statistic.
’s have been defined previously for calculating the Lagrange multiplier statistic.
Essentially, the numerator and denominator estimate the number of failures and the number at risks that are associated with the Turnbull intervals. Thus these quantities estimate the increments of the cumulative hazard function over the Turnbull intervals.
The estimator of the cumulative hazard function is
Like ![]() ,
, ![]() is undefined if t is located within some Turnbull interval
is undefined if t is located within some Turnbull interval ![]() . To facilitate applying the kernel-smoothed methods, you need to reformulate the estimator so that it has only point masses.
An ad hoc approach would be to place all the mass for a Turnbull interval at the right boundary. The kernel-based estimate
of the hazard function is computed as
. To facilitate applying the kernel-smoothed methods, you need to reformulate the estimator so that it has only point masses.
An ad hoc approach would be to place all the mass for a Turnbull interval at the right boundary. The kernel-based estimate
of the hazard function is computed as
where ![]() is a kernel function and
is a kernel function and ![]() is the bandwidth. You can estimate the cumulative hazard function by integrating
is the bandwidth. You can estimate the cumulative hazard function by integrating ![]() with respect to t.
with respect to t.
Practically, an upper limit ![]() is usually imposed so that the kernel-smoothed estimate is defined on
is usually imposed so that the kernel-smoothed estimate is defined on ![]() . The ICLIFETEST procedure sets the value depending on whether the right boundary of the last Turnbull interval is finite
or not:
. The ICLIFETEST procedure sets the value depending on whether the right boundary of the last Turnbull interval is finite
or not: ![]() if
if ![]() and
and ![]() otherwise.
otherwise.
Typical choices of kernel function are as follows:
-
uniform kernel:
![\[ K_ U(x) = \frac{1}{2}, ~ ~ -1\leq x \leq 1\\ \]](images/statug_iclifetest0169.png)
-
Epanechnikov kernel:
![\[ K_ E(x) = \frac{3}{4}(1-x^2), ~ ~ -1\leq x \leq 1 \]](images/statug_iclifetest0170.png)
-
biweight kernel:
![\[ K_{\mi{BW}}(x) = \frac{15}{16}(1-x^2)^2, ~ ~ -1\leq x \leq 1 \]](images/statug_iclifetest0171.png)
For t < b, the symmetric kernels ![]() are replaced by the corresponding asymmetric kernels of Gasser and Müller (1979). Let
are replaced by the corresponding asymmetric kernels of Gasser and Müller (1979). Let ![]() . The modified kernels are as follows:
. The modified kernels are as follows:
-
uniform kernel:
![\[ K_{U,q}(x) = \frac{4(1+q^3)}{(1+q)^4} + \frac{6(1-q)}{(1+q)^3}x, ~ ~ ~ -1 \leq x \leq q \]](images/statug_iclifetest0174.png)
-
Epanechnikov kernel:
![\[ K_{E,q}(x)= K_ E(x) \frac{64(2-4q+6q^2-3q^3) + 240(1-q)^2 x }{ (1+q)^4(19-18q+3q^2)}, ~ ~ -1 \leq x \leq q \]](images/statug_iclifetest0175.png)
-
biweight kernel:
![\[ K_{\mi{BW},q}(x) = K_{\mi{BW}}(x) \frac{64(8-24q+48q^2-45q^3+15q^4) + 1120(1-q)^3 x}{(1+q)^5(81-168q+126q^2-40q^3+5q^4)}, ~ ~ -1\leq x \leq q \]](images/statug_iclifetest0176.png)
For ![]() , let
, let ![]() . The asymmetric kernels for
. The asymmetric kernels for ![]() are used, with x replaced by –x.
are used, with x replaced by –x.
The bandwidth parameter b controls how much “smoothness” you want to have in the kernel-smoothed estimate. For right-censored data, a commonly accepted method of choosing an optimal bandwidth is to use the mean integrated square error(MISE) as an objective criteria. This measure becomes difficult to adapt to interval-censored data because it no longer has a closed-form mathematical formula.
Pan (2000) proposes using a V-fold cross validation likelihood as a criterion for choosing the optimal bandwidth for the kernel-smoothed estimate of the
survival function. The ICLIFETEST procedure implements this approach for smoothing the hazard function. Computing such a criterion
entails a cross validation type procedure. First, the original data ![]() are partitioned into V almost balanced subsets
are partitioned into V almost balanced subsets ![]() ,
, ![]() . Denote the kernel-smoothed estimate of the leave-one-subset-out data
. Denote the kernel-smoothed estimate of the leave-one-subset-out data ![]() as
as ![]() . The optimal bandwidth is defined as the one that maximizes the cross validation likelihood:
. The optimal bandwidth is defined as the one that maximizes the cross validation likelihood:
If the TEST statement is specified, the ICLIFETEST procedure compares the K groups formed by the levels of the TEST variable using a generalized log-rank test. Let ![]() be the underlying survival function of the kth group,
be the underlying survival function of the kth group, ![]() . The null and alternative hypotheses to be tested are
. The null and alternative hypotheses to be tested are
![]() for all t
for all t
versus
![]() at least one of the
at least one of the ![]() ’s is different for some t
’s is different for some t
Let ![]() denote the number of subjects in group k, and let n denote the total number of subjects (
denote the number of subjects in group k, and let n denote the total number of subjects (![]() ).
).
For the ith subject, let ![]() be a vector of K indicators that represent whether or not the subject belongs to the kth group. Denote
be a vector of K indicators that represent whether or not the subject belongs to the kth group. Denote ![]() , where
, where ![]() represents the treatment effect for the kth group. Suppose that a model is specified and the survival function for the ith subject can be written as
represents the treatment effect for the kth group. Suppose that a model is specified and the survival function for the ith subject can be written as
where ![]() denotes the nuisance parameters.
denotes the nuisance parameters.
It follows that the likelihood function is
where ![]() denotes the interval observation for the ith subject.
denotes the interval observation for the ith subject.
Testing whether or not the survival functions are equal across the K groups is equivalent to testing whether all the ![]() ’s are zero. It is natural to consider a score test based on the specified model (Finkelstein, 1986).
’s are zero. It is natural to consider a score test based on the specified model (Finkelstein, 1986).
The score statistics for ![]() are derived as the first-order derivatives of the log-likelihood function evaluated at
are derived as the first-order derivatives of the log-likelihood function evaluated at ![]() and
and ![]() .
.
where ![]() denotes the maximum likelihood estimate for the
denotes the maximum likelihood estimate for the ![]() , given that
, given that ![]() .
.
Under the null hypothesis that ![]() , all K groups share the same survival function
, all K groups share the same survival function ![]() . It is typical to leave
. It is typical to leave ![]() unspecified and obtain a nonparametric maximum likelihood estimate
unspecified and obtain a nonparametric maximum likelihood estimate ![]() using, for instance, Turnbull’s method. In this case,
using, for instance, Turnbull’s method. In this case, ![]() represents all the parameters to be estimated in order to determine
represents all the parameters to be estimated in order to determine ![]() .
.
Suppose the given data generates m Turnbull intervals as ![]() . Denote the probability estimate at the right end point of the jth interval by
. Denote the probability estimate at the right end point of the jth interval by ![]() . The nonparametric survival estimate is
. The nonparametric survival estimate is ![]() for
for ![]() any
any ![]() .
.
Under the null hypothesis, Fay (1999) showed that the score statistics can be written in the form of a weighted log-rank test as
where
and ![]() denotes the derivative of
denotes the derivative of ![]() with respect to
with respect to ![]() .
.
![]() estimates the expected number of events within
estimates the expected number of events within ![]() for the kth group, and it is computed as
for the kth group, and it is computed as
![]() is an estimate for the expected number of events within
is an estimate for the expected number of events within ![]() for the whole sample, and it is computed as
for the whole sample, and it is computed as
Similarly, ![]() estimates the expected number of subjects at risk before entering
estimates the expected number of subjects at risk before entering ![]() for the kth group, and can be estimated by
for the kth group, and can be estimated by ![]() .
. ![]() is an estimate of the expected number of subjects at risk before entering
is an estimate of the expected number of subjects at risk before entering ![]() for all the groups:
for all the groups: ![]() .
.
Assuming different survival models gives rise to different weight functions ![]() (Fay, 1999). For example, Finkelstein’s score test (1986) is derived assuming a proportional hazards model; Fay’s test (1996) is based on a proportional odds model.
(Fay, 1999). For example, Finkelstein’s score test (1986) is derived assuming a proportional hazards model; Fay’s test (1996) is based on a proportional odds model.
The choices of weight function are given in Table 50.3.
Sun (1996) proposed the use of multiple imputation to estimate the variance-covariance matrix of the generalized log-rank statistic
![]() . This approach is similar to the multiple imputation method as presented in Variance Estimation of the Survival Estimator. Both methods impute right-censored data from interval-censored data and analyze the imputed data sets by using standard
statistical techniques. Huang, Lee, and Yu (2008) suggested improving the performance of the generalized log-rank test by slightly modifying the variance calculation.
. This approach is similar to the multiple imputation method as presented in Variance Estimation of the Survival Estimator. Both methods impute right-censored data from interval-censored data and analyze the imputed data sets by using standard
statistical techniques. Huang, Lee, and Yu (2008) suggested improving the performance of the generalized log-rank test by slightly modifying the variance calculation.
Suppose the given data generate m Turnbull intervals as ![]() . Denote the probability estimate for the jth interval as
. Denote the probability estimate for the jth interval as ![]() , and denote the nonparametric survival estimate as
, and denote the nonparametric survival estimate as ![]() for
for ![]() any
any ![]() .
.
In order to generate an imputed data set, you need to randomly generate a survival time for every subject of the sample. For
the ith subject, a random time ![]() is generated randomly based on the following discrete survival function:
is generated randomly based on the following discrete survival function:
where ![]() denotes the interval observation for the subject.
denotes the interval observation for the subject.
For the hth imputed data set (![]() ), let
), let ![]() and
and ![]() denote the numbers of failures and subjects at risk by counting the imputed
denote the numbers of failures and subjects at risk by counting the imputed ![]() ’s for group k. Let
’s for group k. Let ![]() and
and ![]() denote the corresponding pooled numbers.
denote the corresponding pooled numbers.
You can perform the standard weighted log-rank test for right-censored data on each of the imputed data sets (Huang, Lee, and Yu, 2008). The test statistic is
where
Its variance-covariance matrix is estimated by the Greenwood formula as
where
After analyzing each imputed data set, you can estimate the variance-covariance matrix of ![]() by pooling the results as
by pooling the results as
where
The overall test statistic is formed as ![]() , where
, where ![]() is the generalized inverse of
is the generalized inverse of ![]() . Under the null hypothesis, the statistic has a chi-squared distribution with degrees of freedom equal to the rank of
. Under the null hypothesis, the statistic has a chi-squared distribution with degrees of freedom equal to the rank of ![]() . By default, the ICLIFETEST procedure perform 1000 imputations. You can change the number of imputations by the IMPUTE option
in the PROC ICLIFETEST statement.
. By default, the ICLIFETEST procedure perform 1000 imputations. You can change the number of imputations by the IMPUTE option
in the PROC ICLIFETEST statement.
Suppose the generalized log-rank test is to be stratified on the M levels that are formed from the variables that you specify in the STRATA statement. Based only on the data of the sth stratum (![]() ), let
), let ![]() be the test statistic for the sth stratum and let
be the test statistic for the sth stratum and let ![]() be the corresponding covariance matrix as constructed in the section Variance Estimation of the Generalized Log-Rank Statistic. First, sum over the stratum-specific estimates as follows:
be the corresponding covariance matrix as constructed in the section Variance Estimation of the Generalized Log-Rank Statistic. First, sum over the stratum-specific estimates as follows:
Then construct the global test statistic as
Under the null hypothesis, the test statistic has a chi-squared distribution with degrees of freedom equal to the rank of
![]() . The ICLIFETEST procedure performs the stratified test only when the groups to be compared are balanced across all the strata.
. The ICLIFETEST procedure performs the stratified test only when the groups to be compared are balanced across all the strata.
When you have more than two groups, a generalized log-rank test tells you whether the survival curves are significantly different from each other, but it does not identify which pairs of curves are different. Pairwise comparisons can be performed based on the generalized log-rank statistic and the corresponding variance-covariance matrix. However, reporting all pairwise comparisons is problematic because the overall Type I error rate would be inflated. A multiple-comparison adjustment of the p-values for the paired comparisons retains the same overall probability of a Type I error as the K-sample test.
The ICLIFETEST procedure supports two types of paired comparisons: comparisons between all pairs of curves and comparisons between a control curve and all other curves. You use the DIFF= option to specify the comparison type, and you use the ADJUST= option to select a method of multiple-comparison adjustments.
Let ![]() denote a chi-square random variable with r degrees of freedom. Denote
denote a chi-square random variable with r degrees of freedom. Denote ![]() and
and ![]() as the density function and the cumulative distribution function of a standard normal distribution, respectively. Let m be the number of comparisons; that is,
as the density function and the cumulative distribution function of a standard normal distribution, respectively. Let m be the number of comparisons; that is,
For a two-sided test that compares the survival of the jth group with that of lth group, ![]() , the test statistic is
, the test statistic is
and the raw p-value is
For multiple comparisons of more than two groups (![]() ), adjusted p-values are computed as follows:
), adjusted p-values are computed as follows:
-
Bonferroni adjustment:
![\[ p = \mr{min}\{ 1, m \mr{Pr}(\chi ^2_1 > z^2_{jl})\} \]](images/statug_iclifetest0253.png)
-
Dunnett-Hsu adjustment: With the first group defined as the control, there are
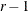 comparisons to be made. Let
comparisons to be made. Let 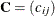 be the
be the 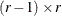 matrix of contrasts that represents the
matrix of contrasts that represents the 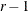 comparisons; that is,
comparisons; that is,
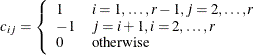
Let
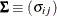 and
and 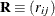 be covariance and correlation matrices of
be covariance and correlation matrices of 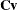 , respectively; that is,
, respectively; that is,
![\[ \bSigma = \bC \bV \bC ’ \]](images/statug_iclifetest0261.png)
and
![\[ r_{ij}= \frac{\sigma _{ij}}{\sqrt {\sigma _{ii}\sigma _{jj}}} \]](images/statug_iclifetest0262.png)
The factor-analytic covariance approximation of Hsu (1992) is to find
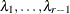 such that
such that
![\[ \bR = \bD + \blambda \blambda ’ \]](images/statug_iclifetest0264.png)
where
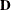 is a diagonal matrix whose jth diagonal element is
is a diagonal matrix whose jth diagonal element is 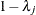 and
and 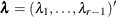 . The adjusted p-value is
. The adjusted p-value is
![\[ p= 1 - \int _{-\infty }^{\infty } \phi (y) \prod _{i=1}^{r-1} \biggl [ \Phi \biggl (\frac{\lambda _ i y + z_{jl}}{\sqrt {1-\lambda _ i^2}}\biggr ) - \Phi \biggl (\frac{\lambda _ i y - z_{jl}}{\sqrt {1-\lambda _ i^2}} \biggr ) \biggr ]dy \]](images/statug_iclifetest0268.png)
This value can be obtained in a DATA step as
![\[ p=\mr{PROBMC}(\mr{"DUNNETT2"}, z_{ij},.,.,r-1,\lambda _1,\ldots ,\lambda _{r-1}). \]](images/statug_iclifetest0269.png)
-
Scheffé adjustment:
![\[ p = \mr{Pr}(\chi ^2_{r-1} > z^2_{jl}) \]](images/statug_iclifetest0270.png)
-
Šidák adjustment:
![\[ p = 1-\{ 1- \mr{Pr}(\chi ^2_1 > z^2_{jl})\} ^ m \]](images/statug_iclifetest0271.png)
-
SMM adjustment:
![\[ p = 1 - [2\Phi (z_{jl}) -1]^ m \]](images/statug_iclifetest0272.png)
This can also be evaluated in a DATA step as
![\[ p = 1 - \mr{PROBMC}(\mr{"MAXMOD"},z_{jl},.,.,m). \]](images/statug_iclifetest0273.png)
-
Tukey adjustment:
![\[ p = 1 - \int _{-\infty }^{\infty } r \phi (y)[\Phi (y) - \Phi (y-\sqrt {2}z_{jl})]^{r-1}dy \]](images/statug_iclifetest0274.png)
This can be evaluated in a DATA step as
![\[ p = 1 - \mr{PROBMC}(\mr{"RANGE"},\sqrt {2}z_{jl},.,.,r). \]](images/statug_iclifetest0275.png)
Trend tests for right-censored data (Klein and Moeschberger, 1997, Section 7.4) can be extended to interval-censored data in a straightforward way. Such tests are specifically designed to detect ordered alternatives as
![]() with at least one inequality
with at least one inequality
or
![]() with at least one inequality
with at least one inequality
Let ![]() be a sequence of scores associated with the k samples. Let
be a sequence of scores associated with the k samples. Let ![]() be the generalized log-rank statistic and
be the generalized log-rank statistic and ![]() be the corresponding covariance matrix of size
be the corresponding covariance matrix of size ![]() as constructed in the section Variance Estimation of the Generalized Log-Rank Statistic. The trend test statistic and its standard error are given by
as constructed in the section Variance Estimation of the Generalized Log-Rank Statistic. The trend test statistic and its standard error are given by ![]() and
and ![]() , respectively. Under the null hypothesis that there is no trend, the following z-score has, asymptotically, a standard normal distribution:
, respectively. Under the null hypothesis that there is no trend, the following z-score has, asymptotically, a standard normal distribution:
The ICLIFETEST procedure provides both one-tail and two-tail p-values for the test.
