The PHREG Procedure
- Overview
-
Getting Started

-
Syntax
 PROC PHREG StatementASSESS StatementBASELINE StatementBAYES StatementBY StatementCLASS StatementCONTRAST StatementEFFECT StatementESTIMATE StatementFREQ StatementHAZARDRATIO StatementID StatementLSMEANS StatementLSMESTIMATE StatementMODEL StatementOUTPUT StatementProgramming StatementsRANDOM StatementSTRATA StatementSLICE StatementSTORE StatementTEST StatementWEIGHT Statement
PROC PHREG StatementASSESS StatementBASELINE StatementBAYES StatementBY StatementCLASS StatementCONTRAST StatementEFFECT StatementESTIMATE StatementFREQ StatementHAZARDRATIO StatementID StatementLSMEANS StatementLSMESTIMATE StatementMODEL StatementOUTPUT StatementProgramming StatementsRANDOM StatementSTRATA StatementSLICE StatementSTORE StatementTEST StatementWEIGHT Statement -
Details
 Failure Time DistributionTime and CLASS Variables UsagePartial Likelihood Function for the Cox ModelCounting Process Style of InputLeft-Truncation of Failure TimesThe Multiplicative Hazards ModelThe Frailty ModelHazard RatiosProportional Rates/Means Models for Recurrent EventsNewton-Raphson MethodFirth’s Modification for Maximum Likelihood EstimationRobust Sandwich Variance EstimateTesting the Global Null HypothesisType 3 TestsConfidence Limits for a Hazard RatioUsing the TEST Statement to Test Linear HypothesesAnalysis of Multivariate Failure Time DataModel Fit StatisticsResidualsDiagnostics Based on Weighted ResidualsInfluence of Observations on Overall Fit of the ModelSurvivor Function EstimatorsEffect Selection MethodsAssessment of the Proportional Hazards ModelThe Penalized Partial Likelihood Approach for Fitting Frailty ModelsSpecifics for Bayesian AnalysisComputational ResourcesInput and Output Data SetsDisplayed OutputODS Table NamesODS Graphics
Failure Time DistributionTime and CLASS Variables UsagePartial Likelihood Function for the Cox ModelCounting Process Style of InputLeft-Truncation of Failure TimesThe Multiplicative Hazards ModelThe Frailty ModelHazard RatiosProportional Rates/Means Models for Recurrent EventsNewton-Raphson MethodFirth’s Modification for Maximum Likelihood EstimationRobust Sandwich Variance EstimateTesting the Global Null HypothesisType 3 TestsConfidence Limits for a Hazard RatioUsing the TEST Statement to Test Linear HypothesesAnalysis of Multivariate Failure Time DataModel Fit StatisticsResidualsDiagnostics Based on Weighted ResidualsInfluence of Observations on Overall Fit of the ModelSurvivor Function EstimatorsEffect Selection MethodsAssessment of the Proportional Hazards ModelThe Penalized Partial Likelihood Approach for Fitting Frailty ModelsSpecifics for Bayesian AnalysisComputational ResourcesInput and Output Data SetsDisplayed OutputODS Table NamesODS Graphics -
Examples
 Stepwise RegressionBest Subset SelectionModeling with Categorical PredictorsFirth’s Correction for Monotone LikelihoodConditional Logistic Regression for m:n MatchingModel Using Time-Dependent Explanatory VariablesTime-Dependent Repeated Measurements of a CovariateSurvival CurvesAnalysis of ResidualsAnalysis of Recurrent Events DataAnalysis of Clustered DataModel Assessment Using Cumulative Sums of Martingale ResidualsBayesian Analysis of the Cox ModelBayesian Analysis of Piecewise Exponential Model
Stepwise RegressionBest Subset SelectionModeling with Categorical PredictorsFirth’s Correction for Monotone LikelihoodConditional Logistic Regression for m:n MatchingModel Using Time-Dependent Explanatory VariablesTime-Dependent Repeated Measurements of a CovariateSurvival CurvesAnalysis of ResidualsAnalysis of Recurrent Events DataAnalysis of Clustered DataModel Assessment Using Cumulative Sums of Martingale ResidualsBayesian Analysis of the Cox ModelBayesian Analysis of Piecewise Exponential Model - References
To request a Bayesian analysis, you specify the new BAYES statement in addition to the PROC PHREG statement and the MODEL statement. You include a CLASS statement if you have effects that involve categorical variables. The FREQ or WEIGHT statement can be included if you have a frequency or weight variable, respectively, in the input data. The STRATA statement can be used to carry out a stratified analysis for the Cox model, but it is not allowed in the piecewise constant baseline hazard model. Programming statements can be used to create time-dependent covariates for the Cox model, but they are not allowed in the piecewise constant baseline hazard model. However, you can use the counting process style of input to accommodate time-dependent covariates that are not continuously changing with time for the piecewise constant baseline hazard model and the Cox model as well. The HAZARDRATIO statement enables you to request a hazard ratio analysis based on the posterior samples. The ASSESS, CONTRAST, ID, OUTPUT, and TEST statements, if specified, are ignored. Also ignored are the COVM and COVS options in the PROC PHREG statement and the following options in the MODEL statement: BEST=, CORRB, COVB, DETAILS, HIERARCHY=, INCLUDE=, MAXSTEP=, NOFIT, PLCONV=, SELECTION=, SEQUENTIAL, SLENTRY=, and SLSTAY=.
Let ![]() be the observed data. Let
be the observed data. Let ![]() be a partition of the time axis.
be a partition of the time axis.
The hazard function for subject i is
where
The baseline cumulative hazard function is
where
 |
The log likelihood is given by
|
|
|
|
|
|
|
|
where ![]() .
.
Note that for ![]() , the full conditional for
, the full conditional for ![]() is log-concave only when
is log-concave only when ![]() , but the full conditionals for the
, but the full conditionals for the ![]() ’s are always log-concave.
’s are always log-concave.
For a given ![]() ,
, ![]() gives
gives
Substituting these values into ![]() gives the profile log likelihood for
gives the profile log likelihood for ![]()
where ![]() . Since the constant c does not depend on
. Since the constant c does not depend on ![]() , it can be discarded from
, it can be discarded from ![]() in the optimization.
in the optimization.
The MLE ![]() of
of ![]() is obtained by maximizing
is obtained by maximizing
with respect to ![]() , and the MLE
, and the MLE ![]() of
of ![]() is given by
is given by
For ![]() , let
, let
|
|
|
|
|
|
|
|
The partial derivatives of ![]() are
are
|
|
|
|
|
|
|
![$\displaystyle \sum _{j=1}^ J d_ j \biggl \{ \frac{\bS _ j^{(2)}(\bbeta )}{S_ j^{(0)}(\bbeta )} - \biggl [\bE _ j(\bbeta )\biggl ] \biggl [ \bE _ j(\bbeta )\biggr ]’\biggl \} $](images/statug_phreg0816.png) |
The asymptotic covariance matrix for ![]() is obtained as the inverse of the information matrix given by
is obtained as the inverse of the information matrix given by
|
|
|
|
|
|
|
|
|
|
|
|
See Example 6.5.1 in Lawless (2003) for details.
By letting
you can build a prior correlation among the ![]() ’s by using a correlated prior
’s by using a correlated prior ![]() , where
, where ![]() .
.
The log likelihood is given by
Then the MLE of ![]() is given by
is given by
Note that the full conditionals for ![]() ’s and
’s and ![]() ’s are always log-concave.
’s are always log-concave.
The asymptotic covariance matrix for ![]() is obtained as the inverse of the information matrix formed by
is obtained as the inverse of the information matrix formed by
|
|
|
|
|
|
|
|
|
|
|
|
For a Cox model, the model parameters are the regression coefficients. For a piecewise exponential model, the model parameters consist of the regression coefficients and the hazards or log-hazards. The priors for the hazards and the priors for the regression coefficients are assumed to be independent, while you can have a joint multivariate normal prior for the log-hazards and the regression coefficients.
Let ![]() be the constant baseline hazards.
be the constant baseline hazards.
The joint prior density is given by
This prior is improper (nonintegrable), but the posterior distribution is proper as long as there is at least one event time in each of the constant hazard intervals.
The joint prior density is given by
This prior is improper (nonintegrable), but the posteriors are proper as long as there is at least one event time in each of the constant hazard intervals.
The gamma distribution ![]() has a PDF
has a PDF
where a is the shape parameter and ![]() is the scale parameter. The mean is
is the scale parameter. The mean is ![]() and the variance is
and the variance is ![]() .
.
Write ![]() .
.
The joint prior density is given by
Note that the uniform prior for the log-hazards is the same as the improper prior for the hazards.
Let ![]() be the vector of regression coefficients.
be the vector of regression coefficients.
The joint prior density is given by
This prior is improper, but the posterior distributions for ![]() are proper.
are proper.
Assume ![]() has a multivariate normal prior with mean vector
has a multivariate normal prior with mean vector ![]() and covariance matrix
and covariance matrix ![]() . The joint prior density is given by
. The joint prior density is given by
Assume ![]() has a multivariate normal prior with mean vector
has a multivariate normal prior with mean vector ![]() and covariance matrix
and covariance matrix ![]() . The joint prior density is given by
. The joint prior density is given by
Assume ![]() has a multivariate normal prior with mean vector
has a multivariate normal prior with mean vector ![]() and covariance matrix
and covariance matrix ![]() , where
, where ![]() is the design matrix and g is either a constant or it follows a gamma prior with density
is the design matrix and g is either a constant or it follows a gamma prior with density ![]() where a and b are the SHAPE= and ISCALE= parameters. Let k be the rank of
where a and b are the SHAPE= and ISCALE= parameters. Let k be the rank of ![]() . The joint prior density with g being a constant c is given by
. The joint prior density with g being a constant c is given by
The joint prior density with g having a gamma prior is given by
Denote the observed data by D.
For the Gibbs sampler, PROC PHREG uses the ARMS (adaptive rejection Metropolis sampling) algorithm of Gilks, Best, and Tan (1995) to sample from the full conditionals. This is the default sampling scheme. Alternatively, you can requests the random walk Metropolis (RWM) algorithm to sample an entire parameter vector from the posterior distribution. For a general discussion of these algorithms, see section Markov Chain Monte Carlo Method.
You can output these posterior samples into a SAS data set by using the OUTPOST= option in the BAYES statement, or you can
use the following SAS statement to output the posterior samples into the SAS data set Post:
ods output PosteriorSample=Post;
The output data set also includes the variables LogLike and LogPost, which represent the log of the likelihood and the log of the posterior log density, respectively.
Let ![]() be the parameter vector. For the Cox model, the
be the parameter vector. For the Cox model, the ![]() ’s are the regression coefficients
’s are the regression coefficients ![]() ’s, and for the piecewise constant baseline hazard model, the
’s, and for the piecewise constant baseline hazard model, the ![]() ’s consist of the baseline hazards
’s consist of the baseline hazards ![]() ’s (or log baseline hazards
’s (or log baseline hazards ![]() ’s) and the regression coefficients
’s) and the regression coefficients ![]() ’s. Let
’s. Let ![]() be the likelihood function, where D is the observed data. Note that for the Cox model, the likelihood contains the infinite-dimensional baseline hazard function,
and the gamma process is perhaps the most commonly used prior process (Ibrahim, Chen, and Sinha, 2001). However, Sinha, Ibrahim, and Chen (2003) justify using the partial likelihood as the likelihood function for the Bayesian analysis. Let
be the likelihood function, where D is the observed data. Note that for the Cox model, the likelihood contains the infinite-dimensional baseline hazard function,
and the gamma process is perhaps the most commonly used prior process (Ibrahim, Chen, and Sinha, 2001). However, Sinha, Ibrahim, and Chen (2003) justify using the partial likelihood as the likelihood function for the Bayesian analysis. Let ![]() be the prior distribution. The posterior
be the prior distribution. The posterior ![]() is proportional to the joint distribution
is proportional to the joint distribution ![]() .
.
The full conditional distribution of ![]() is proportional to the joint distribution; that is,
is proportional to the joint distribution; that is,
For example, the one-dimensional conditional distribution of ![]() , given
, given ![]() , is computed as
, is computed as
Suppose you have a set of arbitrary starting values ![]() . Using the ARMS algorithm, an iteration of the Gibbs sampler consists of the following:
. Using the ARMS algorithm, an iteration of the Gibbs sampler consists of the following:
-
draw
 from
from 
-
draw
 from
from 
-

-
draw
 from
from 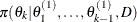
After one iteration, you have ![]() . After n iterations, you have
. After n iterations, you have ![]() . Cumulatively, a chain of n samples is obtained.
. Cumulatively, a chain of n samples is obtained.
PROC PHREG uses a multivariate normal proposal distribution ![]() centered at
centered at ![]() . With an initial parameter vector
. With an initial parameter vector ![]() , a new sample
, a new sample ![]() is obtained as follows:
is obtained as follows:
-
sample
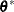 from
from 
-
calculate the quantity

-
sample u from the uniform distribution

-
set
 if
if  ; otherwise set
; otherwise set 
With ![]() taking the role of
taking the role of ![]() , the previous steps are repeated to generate the next sample
, the previous steps are repeated to generate the next sample ![]() . After n iterations, a chain of n samples
. After n iterations, a chain of n samples ![]() is obtained.
is obtained.
When the BAYES statement is specified, PROC PHREG generates one Markov chain that contains the approximate posterior samples
of the model parameters. Additional chains are produced when the Gelman-Rubin diagnostics are requested. Starting values (initial
values) can be specified in the INITIAL= data set in the BAYES statement. If the INITIAL= option is not specified, PROC PHREG
picks its own initial values for the chains based on the maximum likelihood estimates of ![]() and the prior information of
and the prior information of ![]() .
.
Denote ![]() as the integral value of x.
as the integral value of x.
For the first chain that the summary statistics and diagnostics are based on, the initial values are
For subsequent chains, the starting values are picked in two different ways according to the total number of chains specified.
If the total number of chains specified is less than or equal to 10, initial values of the rth chain (![]() ) are given by
) are given by
with the plus sign for odd r and minus sign for even r. If the total number of chains is greater than 10, initial values are picked at random over a wide range of values. Let ![]() be a uniform random number between 0 and 1; the initial value for
be a uniform random number between 0 and 1; the initial value for ![]() is given by
is given by
The ![]() ’s are the regression coefficients
’s are the regression coefficients ![]() ’s, and in the piecewise exponential model, include the log-hazard parameters
’s, and in the piecewise exponential model, include the log-hazard parameters ![]() ’s. For the first chain that the summary statistics and regression diagnostics are based on, the initial values are
’s. For the first chain that the summary statistics and regression diagnostics are based on, the initial values are
If the number of chains requested is less than or equal to 10, initial values for the rth chain (![]() ) are given by
) are given by
with the plus sign for odd r and minus sign for even r. When there are more than 10 chains, the initial value for the ![]() is picked at random over the range
is picked at random over the range ![]() ; that is,
; that is,
where ![]() is a uniform random number between 0 and 1.
is a uniform random number between 0 and 1.
Denote the observed data by D. Let ![]() be the vector of parameters of length k. Let
be the vector of parameters of length k. Let ![]() be the likelihood. The deviance information criterion (DIC) proposed in Spiegelhalter et al. (2002) is a Bayesian model assessment tool. Let Dev
be the likelihood. The deviance information criterion (DIC) proposed in Spiegelhalter et al. (2002) is a Bayesian model assessment tool. Let Dev![]() . Let
. Let ![]() and
and ![]() be the corresponding posterior means of
be the corresponding posterior means of ![]() and
and ![]() , respectively. The deviance information criterion is computed as
, respectively. The deviance information criterion is computed as
Also computed is
where pD is interpreted as the effective number of parameters.
Note that ![]() defined here does not have the standardizing term as in the section Deviance Information Criterion (DIC). Nevertheless, the DIC calculated here is still useful for variable selection.
defined here does not have the standardizing term as in the section Deviance Information Criterion (DIC). Nevertheless, the DIC calculated here is still useful for variable selection.
Let ![]() be the parameter vector. For the Cox model, the
be the parameter vector. For the Cox model, the ![]() ’s are the regression coefficients
’s are the regression coefficients ![]() ’s; for the piecewise constant baseline hazard model, the
’s; for the piecewise constant baseline hazard model, the ![]() ’s consist of the baseline hazards
’s consist of the baseline hazards ![]() ’s (or log baseline hazards
’s (or log baseline hazards ![]() ’s) and the regression coefficients
’s) and the regression coefficients ![]() ’s. Let
’s. Let ![]() be the chain that represents the posterior distribution for
be the chain that represents the posterior distribution for ![]() .
.
Consider a quantity of interest ![]() that can be expressed as a function
that can be expressed as a function ![]() of the parameter vector
of the parameter vector ![]() . You can construct the posterior distribution of
. You can construct the posterior distribution of ![]() by evaluating the function
by evaluating the function ![]() for each
for each ![]() in
in ![]() . The posterior chain for
. The posterior chain for ![]() is
is ![]() Summary statistics such as mean, standard deviation, percentiles, and credible intervals are used to describe the posterior
distribution of
Summary statistics such as mean, standard deviation, percentiles, and credible intervals are used to describe the posterior
distribution of ![]() .
.
As shown in the section Hazard Ratios, a log-hazard ratio is a linear combination of the regression coefficients. Let ![]() be the vector of linear coefficients. The posterior sample for this hazard ratio is the set
be the vector of linear coefficients. The posterior sample for this hazard ratio is the set ![]() .
.
Let ![]() be a covariate vector of interest.
be a covariate vector of interest.
Let ![]() be the observed data. Define
be the observed data. Define
|
|
Consider the rth draw ![]() of
of ![]() . The baseline cumulative hazard function at time t is given by
. The baseline cumulative hazard function at time t is given by
For the given covariate vector ![]() , the cumulative hazard function at time t is
, the cumulative hazard function at time t is
and the survival function at time t is
![$\displaystyle \Delta _ j((s_ i,t_ i]) = \left\{ \begin{array}{ll} 0 & t_ i<a_{j-1} \vee s_ i> a_ j \\ t_ i-\max (s_ i,a_{j-1}) & a_{j-1}\le t_ i < a_ j \\ a_ j - \max (s_ i,a_{j-1}) & t_ i \ge a_ j \end{array} \right. $](images/statug_phreg0843.png)
![\[ \pi (\bbeta |D) \propto \underbrace{L_(D|\bbeta )}_{\mr {partial~ likelihood}} \overbrace{p(\bbeta )}^{\mr {prior}} \]](images/statug_phreg0886.png)
![\[ \pi (\bbeta ,\bgamma ,\theta |D) \propto \underbrace{L(D|\bbeta ,\bgamma )}_{\mr {partial~ likelihood}} \overbrace{g(\bgamma |\theta )}^{\mr {random~ effects}} \underbrace{p(\bbeta ) p(\theta )}_{\mr {priors}} \]](images/statug_phreg0887.png)