The MULTTEST Procedure
Example 60.1 Cochran-Armitage Test with Permutation Resampling
This example, from Keith Soper at Merck, illustrates the exact permutation Cochran-Armitage test carried out on permutation resamples. In the following data set, each observation represents an animal. The binary variables S1 and S2 indicate two tumor types, with 0s indicating no tumor (failure) and 1 indicating a tumor (success); note that they have perfect negative association. The grouping variable is Dose.
data a; input S1 S2 Dose @@; datalines; 0 1 1 1 0 1 0 1 1 0 1 1 0 1 1 1 0 1 1 0 2 1 0 2 0 1 2 1 0 2 0 1 2 1 0 2 1 0 3 1 0 3 1 0 3 0 1 3 0 1 3 1 0 3 ;
proc multtest data=a permutation nsample=10000 seed=36607 outperm=pmt; test ca(S1 S2 / permutation=10 uppertailed); class Dose; contrast 'CA Linear Trend' 0 1 2; run; proc print data=pmt; run;
The PROC MULTTEST statement requests 10,000 permutation resamples. The OUTPERM= option creates an output SAS data set pmt used for the exact permutation distribution computed for the CA test.
The TEST statement specifies an upper-tailed Cochran-Armitage linear trend test for S1 and S2. The cutoff for exact permutation calculations is 10, as specified with the PERMUTATION= option in the TEST statement. Since S1 and S2 have 10 and 8 successes, respectively, PROC MULTTEST uses exact permutation distributions to compute the p-values for both variables.
The groups for the CA test are the levels of Dose from the CLASS statement. The trend coefficients applied to these groups are 0, 1, and 2, respectively, as specified in the CONTRAST statement.
Finally, the PROC PRINT statement displays the SAS data set pmt, which contains the permutation distributions.
The results from this analysis are displayed in Output 60.1.1 through Output 60.1.5. You should check the "Model Information" table to verify that the analysis specifications are correct.
| Model Information | |
|---|---|
| Test for discrete variables | Cochran-Armitage |
| Exact permutation distribution used | Everywhere |
| Tails for discrete tests | Upper-tailed |
| Strata weights | None |
| P-value adjustment | Permutation |
| Number of resamples | 10000 |
| Seed | 36607 |
The label and coefficients from the CONTRAST statement are shown in Output 60.1.2.
| Contrast Coefficients | |||
|---|---|---|---|
| Contrast | Dose | ||
| 1 | 2 | 3 | |
| CA Linear Trend | 0 | 1 | 2 |
Output 60.1.3 displays summary statistics for the two test variables, S1 and S2. The Count column lists the number of successes for each level of the CLASS variable, Dose. The NumObs column lists the sample size, and the Percent column lists the percentage of successes in the sample.
| Discrete Variable Tabulations | ||||
|---|---|---|---|---|
| Variable | Dose | Count | NumObs | Percent |
| S1 | 1 | 2 | 6 | 33.33 |
| S1 | 2 | 4 | 6 | 66.67 |
| S1 | 3 | 4 | 6 | 66.67 |
| S2 | 1 | 4 | 6 | 66.67 |
| S2 | 2 | 2 | 6 | 33.33 |
| S2 | 3 | 2 | 6 | 33.33 |
The Raw column in Output 60.1.4 contains the p-values from the CA test, and the Permutation column contains the permutation-adjusted p-values.
| p-Values | |||
|---|---|---|---|
| Variable | Contrast | Raw | Permutation |
| S1 | CA Linear Trend | 0.1993 | 0.4009 |
| S2 | CA Linear Trend | 0.9220 | 1.0000 |
This table shows that, for S1, the adjusted p-value is approximately twice the raw p-value. In fact, resamples with small (large) p-values for S1 have large (small) p-values for S2 due to the perfect negative association of the variables, and hence the permutation-adjusted p-value for S1 should be 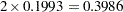 ; the difference is due to resampling error. For the same reason, since the raw p-value for S2 is 0.9220, the adjusted p-value equals 1. The permutation p-values for S1 and S2 also happen to be the Bonferroni-adjusted p-values for this example.
; the difference is due to resampling error. For the same reason, since the raw p-value for S2 is 0.9220, the adjusted p-value equals 1. The permutation p-values for S1 and S2 also happen to be the Bonferroni-adjusted p-values for this example.
The OUTPERM= data set is displayed in Output 60.1.5, which contains the exact permutation distributions for S1 and S2 in terms of cumulative probabilities.
| Obs | _contrast_ | _var_ | _value_ | upper_p |
|---|---|---|---|---|
| 1 | CA Linear Trend | S1 | 0 | 1.00000 |
| 2 | CA Linear Trend | S1 | 1 | 1.00000 |
| 3 | CA Linear Trend | S1 | 2 | 1.00000 |
| 4 | CA Linear Trend | S1 | 3 | 1.00000 |
| 5 | CA Linear Trend | S1 | 4 | 1.00000 |
| 6 | CA Linear Trend | S1 | 5 | 0.99966 |
| 7 | CA Linear Trend | S1 | 6 | 0.99609 |
| 8 | CA Linear Trend | S1 | 7 | 0.97827 |
| 9 | CA Linear Trend | S1 | 8 | 0.92205 |
| 10 | CA Linear Trend | S1 | 9 | 0.80070 |
| 11 | CA Linear Trend | S1 | 10 | 0.61011 |
| 12 | CA Linear Trend | S1 | 11 | 0.38989 |
| 13 | CA Linear Trend | S1 | 12 | 0.19930 |
| 14 | CA Linear Trend | S1 | 13 | 0.07795 |
| 15 | CA Linear Trend | S1 | 14 | 0.02173 |
| 16 | CA Linear Trend | S1 | 15 | 0.00391 |
| 17 | CA Linear Trend | S1 | 16 | 0.00034 |
| 18 | CA Linear Trend | S1 | 17 | 0.00000 |
| 19 | CA Linear Trend | S1 | 18 | 0.00000 |
| 20 | CA Linear Trend | S1 | 19 | 0.00000 |
| 21 | CA Linear Trend | S1 | 20 | 0.00000 |
| 22 | CA Linear Trend | S2 | 0 | 1.00000 |
| 23 | CA Linear Trend | S2 | 1 | 1.00000 |
| 24 | CA Linear Trend | S2 | 2 | 1.00000 |
| 25 | CA Linear Trend | S2 | 3 | 0.99966 |
| 26 | CA Linear Trend | S2 | 4 | 0.99609 |
| 27 | CA Linear Trend | S2 | 5 | 0.97827 |
| 28 | CA Linear Trend | S2 | 6 | 0.92205 |
| 29 | CA Linear Trend | S2 | 7 | 0.80070 |
| 30 | CA Linear Trend | S2 | 8 | 0.61011 |
| 31 | CA Linear Trend | S2 | 9 | 0.38989 |
| 32 | CA Linear Trend | S2 | 10 | 0.19930 |
| 33 | CA Linear Trend | S2 | 11 | 0.07795 |
| 34 | CA Linear Trend | S2 | 12 | 0.02173 |
| 35 | CA Linear Trend | S2 | 13 | 0.00391 |
| 36 | CA Linear Trend | S2 | 14 | 0.00034 |
| 37 | CA Linear Trend | S2 | 15 | 0.00000 |
| 38 | CA Linear Trend | S2 | 16 | 0.00000 |
