| The PRINQUAL Procedure |
| PROC PRINQUAL Statement |
The PROC PRINQUAL statement invokes the PRINQUAL procedure. Optionally, this statement identifies an input data set, creates an output data set, specifies the algorithm and other computational details, and controls displayed output. The options listed in Table 71.1 are available in the PROC PRINQUAL statement.
Option |
Description |
|---|---|
Input Data Set Options |
|
specifies input SAS data set |
|
Output Data Set Details |
|
outputs approximations to transformed variables |
|
specifies prefix for approximation variables |
|
outputs correlations and component structure matrix |
|
specifies a multidimensional preference analysis |
|
specifies output data set |
|
specifies prefix for principal component scores |
|
replaces raw data with transformed data |
|
outputs principal component scores |
|
standardizes principal component scores |
|
specifies prefix for transformed variables |
|
specifies transformation standardization |
|
Method and Iterations |
|
specifies minimum criterion change |
|
specifies number of first iteration to be displayed |
|
specifies minimum data change |
|
analyzes covariances |
|
initializes using dummy variables |
|
specifies number of MAC initialization iterations |
|
specifies maximum number of iterations |
|
specifies iterative algorithm |
|
suppresses numerical error checking |
|
specifies number of principal components |
|
specifies number of MGV models before refreshing |
|
restarts iterations |
|
specifies singularity criterion |
|
specifies input observation type |
|
Missing Data Handling |
|
includes monotone special missing values |
|
excludes observations with missing values |
|
unties special missing values |
|
Control Displayed Output |
|
suppresses displayed output |
|
specifies ODS Graphics details |
|
The following list describes these options in alphabetical order.
- APREFIX=name
- APR=name
specifies a prefix for naming the approximation variables. By default, APREFIX=A. Specifying the APREFIX= option also implies the APPROXIMATIONS option.
- APPROXIMATIONS
- APPROX
- APP
includes principal component approximations to the transformed variables (Eckart and Young; 1936) in the output data set. Variable names are constructed from the value of the APREFIX= option and the input variable names. If you specify the APREFIX= option, then approximations are automatically included. If you specify the APPROXIMATIONS option and not the APREFIX= option, then the APPROXIMATIONS option uses the default, APREFIX=A, to construct the variable names.
- CCONVERGE=n
- CCO=n
specifies the minimum change in the criterion being optimized that is required to continue iterating. By default, CCONVERGE=0.0. The CCONVERGE= option is ignored for METHOD=MAC. For the MGV method, specify CCONVERGE=–2 to ensure data convergence.
- CHANGE=n
- CHA=n
specifies the number of the first iteration to be displayed in the iteration history table. The default is CHANGE=1. When you specify a larger value for
 , the first
, the first 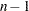 iterations are not displayed, thus speeding up the analysis. The CHANGE= option is most useful with the MGV method, which is much slower than the other methods.
iterations are not displayed, thus speeding up the analysis. The CHANGE= option is most useful with the MGV method, which is much slower than the other methods. - CONVERGE=n
- CON=n
specifies the minimum average absolute change in standardized variable scores that is required to continue iterating. By default, CONVERGE=0.00001. Average change is computed over only those variables that can be transformed by the iterations—that is, all LINEAR, OPSCORE, MONOTONE, UNTIE, SPLINE, MSPLINE, and SSPLINE variables and nonoptimal transformation variables with missing values. For more information, see the section Optimal Transformations.
- COVARIANCE
- COV
computes the principal components from the covariance matrix. The variables are always centered to mean zero. If you do not specify the COVARIANCE option, the variables are also standardized to variance one, which means the analysis is based on the correlation matrix.
- CORRELATIONS
- COR
includes correlations and the component structure matrix in the output data set. By default, this information is not included.
- DATA=SAS-data-set
specifies the SAS data set to be analyzed. The data set must be an ordinary SAS data set; it cannot be a TYPE=CORR or TYPE=COV data set. If you omit the DATA= option, PROC PRINQUAL uses the most recently created SAS data set.
- DUMMY
- DUM
expands variables specified for OPSCORE optimal transformations to dummy variables for the initialization (Tenenhaus and Vachette; 1977). By default, the initial values of OPSCORE variables are the actual data values. The dummy variable nominal initialization requires considerable time and memory, so it might not be possible to use the DUMMY option with large data sets. No separate report of the initialization is produced. Initialization results are incorporated into the first iteration displayed in the iteration history table. For details, see the section Optimal Transformations.
- INITITER=n
- INI=n
specifies the number of MAC iterations required to initialize the data before starting MTV or MGV iterations. By default, INITITER=0. The INITITER= option is ignored if METHOD=MAC.
- MAXITER=n
- MAX=n
specifies the maximum number of iterations. By default, MAXITER=30.
- MDPREF<=n>
- MDP<=n>
specifies a multidimensional preference analysis by implying the STANDARD, SCORES, and CORRELATIONS options. This option also suppresses warnings when there are more variables than observations.
When ODS Graphics is enabled, an MDPREF plot is produced with points for each row and vectors for each column. Often, the vectors are short, and a better graphical display is produced when the vectors are stretched. The absolute lengths of each vector can optionally be changed by specifying MDPREF=
 . Then the vector coordinates are all multiplied by
. Then the vector coordinates are all multiplied by  . Usually,
. Usually,  will be a value such as 2, 2.5, or 3. The default is 2.5. Specify MDPREF=1 to see the vectors without any stretching. The relative lengths of the different vectors is important and interpretable, and these are preserved by the stretching.
will be a value such as 2, 2.5, or 3. The default is 2.5. Specify MDPREF=1 to see the vectors without any stretching. The relative lengths of the different vectors is important and interpretable, and these are preserved by the stretching. - METHOD=MAC | MGV | MTV
- MET=MAC | MGV | MTV
specifies the optimization method. By default, METHOD=MTV. Values of the METHOD= option are MTV, for maximum total variance; MGV, for minimum generalized variance; and MAC, for maximum average correlation. You can use the MAC method when all variables are positively correlated or when no monotonicity constraints are placed on any transformations. See the section The Three Methods of Variable Transformation for more information.
- MONOTONE=two-letters
- MON=two-letters
specifies the first and last special missing value in the list of those special missing values to be estimated using within-variable order and category constraints. By default, there are no order constraints on missing value estimates. The two-letters value must consist of two letters in alphabetical order. For example, MONOTONE=DF means that the estimate of .D must be less than or equal to the estimate of .E, which must be less than or equal to the estimate of .F; no order constraints are placed on estimates of ._, .A through .C, and .G through .Z. For details, see the sections Missing Values and Optimal Scaling in Chapter 91, The TRANSREG Procedure.
- N=n
specifies the number of principal components to be computed. By default, N=2.
- NOCHECK
- NOC
turns off computationally intensive numerical error checking for the MGV method. If you do not specify the NOCHECK option, the procedure computes R square from the squared length of the predicted values vector and compares this value to the R square computed from the error sum of squares that is a byproduct of the sweep algorithm (Goodnight; 1978). If the two values of R square differ by more than the square root of the value of the SINGULAR= option, a warning is displayed, the value of the REFRESH= option is halved, and the model is refit after refreshing. Specifying the NOCHECK option slightly speeds up the algorithm. Note that other less computationally intensive error checking is always performed.
- NOMISS
- NOM
excludes all observations with missing values from the analysis, but does not exclude them from the OUT= data set. If you omit the NOMISS option, PROC PRINQUAL simultaneously computes the optimal transformations of the nonmissing values and estimates the missing values that minimize squared error.
Casewise deletion of observations with missing values occurs when you specify the NOMISS option, when there are missing values in IDENTITY variables, when there are weights less than or equal to 0, or when there are frequencies less than 1. Excluded observations are output with a blank value for the _TYPE_ variable, and they have a weight of 0. They do not contribute to the analysis but are scored and transformed as supplementary or passive observations. See the sections Passive Observations and Missing Values for more information about excluded observations and missing data.
- NOPRINT
- NOP
suppresses the display of all output. This option disables the Output Delivery System (ODS), including ODS Graphics, for the duration of the procedure. For more information, see Chapter 20, Using the Output Delivery System.
- OUT=SAS-data-set
specifies an output SAS data set that contains results of the analysis. If you omit the OUT= option, PROC PRINQUAL still creates an output data set and names it by using the DATA
 convention. If you want to create a permanent SAS data set, you must specify a two-level name. (See the discussion in SAS Language Reference: Concepts.) You can use the REPLACE, APPROXIMATIONS, SCORES, and CORRELATIONS options to control what information is included in the output data set. For details, see the section Output Data Set.
convention. If you want to create a permanent SAS data set, you must specify a two-level name. (See the discussion in SAS Language Reference: Concepts.) You can use the REPLACE, APPROXIMATIONS, SCORES, and CORRELATIONS options to control what information is included in the output data set. For details, see the section Output Data Set. - PLOTS <(global-plot-options)> <= plot-request <(options)>>
- PLOTS <(global-plot-options)> <= (plot-request <(options)> <... plot-request <(options)>>)>
controls the plots produced through ODS Graphics. When you specify only one plot request, you can omit the parentheses from around the plot request. Here are some examples:
plots=none plots=transformation plots(unpack)=transformation
You must enable ODS Graphics before requesting plots, for example, like this:
ods graphics on; proc prinqual plots=all; transformation spline(x1-x10); run; ods graphics off;
For general information about ODS Graphics, see Chapter 21, Statistical Graphics Using ODS. If you have enabled ODS Graphics but do not specify the PLOTS= option, then PROC PRINQUAL produces an MDPREF plot when the MDPREF option is specified.
The global plot options include the following:
- FLIP
- FLI
flips or interchanges the X-axis and Y-axis dimensions for MDPREF plots. The FLIP option can be specified either as a global plot option (for example, PLOTS(FLIP)) or with the MDPREF option (for example, PLOTS=MDPREF(FLIP)).
- INTERPOLATE
- INT
uses observations that are excluded from the analysis for interpolation in the fit and transformation plots. By default, observations with zero weight are excluded from all plots. These include observations with a zero, negative, or missing weight or frequency and observations excluded due to missing and invalid values. You can specify PLOTS(INTERPOLATE)=(plot-requests) to include some of these observations in the plots. You can use this option, for example, with sparse data sets to show smoother functions over the range of the data (see the section The PLOTS(INTERPOLATE) Option in Chapter 91, The TRANSREG Procedure ).
- ONLY
- ONL
suppresses the default plots. Only plots specifically requested are displayed.
- UNPACKPANEL
- UNPACK
- UNP
suppresses paneling. By default, multiple plots can appear in some output panels. Specify UNPACKPANEL to get each plot in a separate panel.
The plot requests include the following:
- ALL
produces all appropriate plots.
- TRANSFORMATION
- TRA
- TRANSFORMATION(UNPACK)
- TRA(UNP)
plots the variable transformations. By default, multiple plots can appear in some output panels. Specify UNPACKPANEL to display each plot in a separate panel.
- MDPREF
- MDP
plots multidimensional preference analysis results. The MDPREF plot can also be requested by specifying the MDPREF option in the PROC statement outside the PLOTS= option.
- NONE
suppresses all plots.
- PREFIX=name
- PRE=name
specifies a prefix for naming the principal components. By default, PREFIX=Prin. As a result, the principal component default names are Prin1, Prin2,..., Prin
 .
. - REFRESH=n
- REF=n
specifies the number of variables to scale in the MGV method before computing a new inverse. By default, REFRESH=5. PROC PRINQUAL uses the REFRESH= option in the sweep algorithm of the MGV method. Large values for the REFRESH= option make the method run faster but with more numerical error. Small values make the method run more slowly but with more numerical accuracy.
- REITERATE
- REI
enables PROC PRINQUAL to use previous transformations as starting points. The REITERATE option affects only variables that are iteratively transformed (specified as LINEAR, SPLINE, MSPLINE, SSPLINE, UNTIE, OPSCORE, and MONOTONE). For iterative transformations, the REITERATE option requests a search in the input data set for a variable that consists of the value of the TPREFIX= option followed by the original variable name. If such a variable is found, it is used to provide the initial values for the first iteration. The final transformation is a member of the transformation family defined by the original variable, not the transformation family defined by the initialization variable. See the section REITERATE Option Usage.
- REPLACE
- REP
replaces the original data with the transformed data in the output data set. The names of the transformed variables in the output data set correspond to the names of the original variables in the input data set. If you do not specify the REPLACE option, both original variables and transformed variables (with names constructed from the TPREFIX= option and the original variable names) are included in the output data set.
- SCORES
- SCO
includes principal component scores in the output data set. By default, scores are not included.
- SINGULAR=n
- SIN=n
specifies the largest value within rounding error of zero. By default, SINGULAR=1E–8. PROC PRINQUAL uses the value of the SINGULAR= option for checking
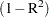 when constructing full-rank matrices of predictor variables, checking denominators before dividing, and so on.
when constructing full-rank matrices of predictor variables, checking denominators before dividing, and so on. - STANDARD
- STD
standardizes the principal component scores in the output data set to mean zero and variance one instead of the default mean zero and variance equal to the corresponding eigenvalue. See the SCORES option.
- TPREFIX=name
- TPR=name
specifies a prefix for naming the transformed variables. By default, TPREFIX=T. The TPREFIX= option is ignored if you specify the REPLACE option.
- TSTANDARD=CENTER | NOMISS | ORIGINAL | Z
- TST=CEN | NOM | ORI | Z
specifies the standardization of the transformed variables in the OUT= data set. By default, TSTANDARD=ORIGINAL. When you specify the TSTANDARD= option in the PROC statement, it the default standardization for all variables. When you specify TSTANDARD= as a t-option, it overrides the default standardization just for selected variables.
- CENTER
centers the output variables to mean zero, but the variances are the same as the variances of the input variables.
- NOMISS
sets the means and variances of the transformed variables in the OUT= data set, computed over all output values that correspond to nonmissing values in the input data set, to the means and variances computed from the nonmissing observations of the original variables. The TSTANDARD=NOMISS specification is useful with missing data. When a variable is linearly transformed, the final variable contains the original nonmissing values and the missing value estimates. In other words, the nonmissing values are unchanged. If your data have no missing values, TSTANDARD=NOMISS and TSTANDARD=ORIGINAL produce the same results.
- ORIGINAL
sets the means and variances of the transformed variables to the means and variances of the original variables. This is the default.
- Z
standardizes the variables to mean zero, variance one.
For nonoptimal variable transformations, the means and variances of the original variables are actually the means and variances of the nonlinearly transformed variables, unless you specify the ORIGINAL nonoptimal t-option in the TRANSFORM statement. For example, if a variable X with no missing values is specified as LOG, then, by default, the final transformation of X is simply LOG(X), not LOG(X) standardized to the mean of X and variance of X.
- TYPE=’text ’|name
- TYP=’text ’|name
specifies the valid value for the _TYPE_ variable in the input data set. If PROC PRINQUAL finds an input _TYPE_ variable, it uses only observations with a _TYPE_ value that matches the TYPE= value. This enables a PROC PRINQUAL OUT= data set containing correlations to be used as input to PROC PRINQUAL without requiring a WHERE statement to exclude the correlations. If a _TYPE_ variable is not in the data set, all observations are used. The default is TYPE=’SCORE’, so if you do not specify the TYPE= option, only observations with _TYPE_ = ’SCORE’ are used.
PROC PRINQUAL displays a note when it reads observations with blank values of _TYPE_, but it does not automatically exclude those observations. Data sets created by the TRANSREG and PRINQUAL procedures have blank _TYPE_ values for those observations that were excluded from the analysis due to nonpositive weights, nonpositive frequencies, or missing data. When these observations are read again, they are excluded for the same reason that they were excluded from their original analysis, not because their _TYPE_ value is blank.
- UNTIE=two-letters
- UNT=two-letters
specifies the first and last special missing values in the list of those special missing values that are to be estimated with within-variable order constraints but no category constraints. The two-letters value must consist of two letters in alphabetical order. By default, there are category constraints but no order constraints on special missing value estimates. For details, see the section Missing Values. Also, see the section Optimal Scaling in Chapter 91, The TRANSREG Procedure.
Copyright © SAS Institute, Inc. All Rights Reserved.