| The GLIMMIX Procedure |
Example 38.4 Quasi-likelihood Estimation for Proportions with Unknown Distribution
Wedderburn (1974) analyzes data on the incidence of leaf blotch (Rhynchosporium secalis) on barley. The data represent the percentage of leaf area affected in a two-way layout with 10 barley varieties at nine sites. The following DATA step converts these data to proportions, as analyzed in McCullagh and Nelder (1989, Ch. 9.2.4). The purpose of the analysis is to make comparisons among the varieties, adjusted for site effects.
data blotch;
array p{9} pct1-pct9;
input variety pct1-pct9;
do site = 1 to 9;
prop = p{site}/100;
output;
end;
drop pct1-pct9;
datalines;
1 0.05 0.00 1.25 2.50 5.50 1.00 5.00 5.00 17.50
2 0.00 0.05 1.25 0.50 1.00 5.00 0.10 10.00 25.00
3 0.00 0.05 2.50 0.01 6.00 5.00 5.00 5.00 42.50
4 0.10 0.30 16.60 3.00 1.10 5.00 5.00 5.00 50.00
5 0.25 0.75 2.50 2.50 2.50 5.00 50.00 25.00 37.50
6 0.05 0.30 2.50 0.01 8.00 5.00 10.00 75.00 95.00
7 0.50 3.00 0.00 25.00 16.50 10.00 50.00 50.00 62.50
8 1.30 7.50 20.00 55.00 29.50 5.00 25.00 75.00 95.00
9 1.50 1.00 37.50 5.00 20.00 50.00 50.00 75.00 95.00
10 1.50 12.70 26.25 40.00 43.50 75.00 75.00 75.00 95.00
;
Little is known about the distribution of the leaf area proportions. The outcomes are not binomial proportions, because they do not represent the ratio of a count over a total number of Bernoulli trials. However, because the mean proportion 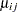 for variety
for variety 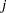 on site
on site 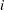 must lie in the interval
must lie in the interval 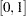 , you can commence the analysis with a model that treats Prop as a "pseudo-binomial" variable:
, you can commence the analysis with a model that treats Prop as a "pseudo-binomial" variable:
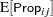 |
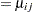 |
|||
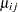 |
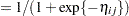 |
|||
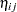 |
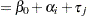 |
|||
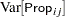 |
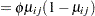 |
Here, 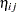 is the linear predictor for variety
is the linear predictor for variety 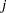 on site
on site 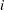 ,
, 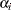 denotes the
denotes the 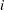 th site effect, and
th site effect, and 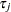 denotes the
denotes the 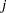 th barley variety effect. The logit of the expected leaf area proportions is linearly related to these effects. The variance funcion of the model is that of a binomial(
th barley variety effect. The logit of the expected leaf area proportions is linearly related to these effects. The variance funcion of the model is that of a binomial( ,
,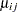 ) variable, and
) variable, and 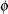 is an overdispersion parameter. The moniker "pseudo-binomial" derives not from the pseudo-likelihood methods used to estimate the parameters in the model, but from treating the response variable as if it had first and second moment properties akin to a binomial random variable.
is an overdispersion parameter. The moniker "pseudo-binomial" derives not from the pseudo-likelihood methods used to estimate the parameters in the model, but from treating the response variable as if it had first and second moment properties akin to a binomial random variable.
The model is fit in the GLIMMIX procedure with the following statements:
proc glimmix data=blotch;
class site variety;
model prop = site variety / link=logit dist=binomial;
random _residual_;
lsmeans variety / diff=control('1');
run;
The MODEL statement specifies the distribution as binomial and the logit link. Because the variance function of the binomial distribution is  , you use the statement
, you use the statement
random _residual_;
to specify the scale parameter 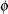 . The LSMEANS statement requests estimates of the least squares means for the barley variety. The DIFF=CONTROL(’1’) option requests tests of least squares means differences against the first variety.
. The LSMEANS statement requests estimates of the least squares means for the barley variety. The DIFF=CONTROL(’1’) option requests tests of least squares means differences against the first variety.
The "Model Information" table in Output 38.4.1 describes the model and methods used in fitting the statistical model. It is assumed here that the data are binomial proportions.
The "Class Level Information" table in Output 38.4.2 lists the number of levels of the Site and Variety effects and their values. All 90 observations read from the data are used in the analysis.
In Output 38.4.3, the "Dimensions" table shows that the model does not contain G-side random effects. There is a single covariance parameter, which corresponds to 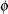 . The "Optimization Information" table shows that the optimization comprises 18 parameters (Output 38.4.3). These correspond to the 18 nonsingular columns of the
. The "Optimization Information" table shows that the optimization comprises 18 parameters (Output 38.4.3). These correspond to the 18 nonsingular columns of the 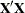 matrix.
matrix.
There are significant site and variety effects in this model based on the approximate Type III 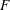 tests (Output 38.4.4).
tests (Output 38.4.4).
Output 38.4.5 displays the Variety least squares means for this analysis. These are obtained by averaging
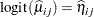 |
across the sites. In other words, LS-means are computed on the linked scale where the model effects are additive. Note that the least squares means are ordered by variety. The estimate of the expected proportion of infected leaf area for the first variety is
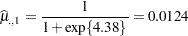 |
and that for the last variety is
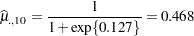 |
| variety Least Squares Means | |||||
|---|---|---|---|---|---|
| variety | Estimate | Standard Error | DF | t Value | Pr > |t| |
| 1 | -4.3800 | 0.5643 | 72 | -7.76 | <.0001 |
| 2 | -4.2300 | 0.5383 | 72 | -7.86 | <.0001 |
| 3 | -3.6906 | 0.4623 | 72 | -7.98 | <.0001 |
| 4 | -3.3319 | 0.4239 | 72 | -7.86 | <.0001 |
| 5 | -2.7653 | 0.3768 | 72 | -7.34 | <.0001 |
| 6 | -2.0089 | 0.3320 | 72 | -6.05 | <.0001 |
| 7 | -1.8095 | 0.3228 | 72 | -5.61 | <.0001 |
| 8 | -1.0380 | 0.2960 | 72 | -3.51 | 0.0008 |
| 9 | -0.8800 | 0.2921 | 72 | -3.01 | 0.0036 |
| 10 | -0.1270 | 0.2808 | 72 | -0.45 | 0.6523 |
Because of the ordering of the least squares means, the differences against the first variety are also ordered from smallest to largest (Output 38.4.6).
| Differences of variety Least Squares Means | ||||||
|---|---|---|---|---|---|---|
| variety | _variety | Estimate | Standard Error | DF | t Value | Pr > |t| |
| 2 | 1 | 0.1501 | 0.7237 | 72 | 0.21 | 0.8363 |
| 3 | 1 | 0.6895 | 0.6724 | 72 | 1.03 | 0.3086 |
| 4 | 1 | 1.0482 | 0.6494 | 72 | 1.61 | 0.1109 |
| 5 | 1 | 1.6147 | 0.6257 | 72 | 2.58 | 0.0119 |
| 6 | 1 | 2.3712 | 0.6090 | 72 | 3.89 | 0.0002 |
| 7 | 1 | 2.5705 | 0.6065 | 72 | 4.24 | <.0001 |
| 8 | 1 | 3.3420 | 0.6015 | 72 | 5.56 | <.0001 |
| 9 | 1 | 3.5000 | 0.6013 | 72 | 5.82 | <.0001 |
| 10 | 1 | 4.2530 | 0.6042 | 72 | 7.04 | <.0001 |
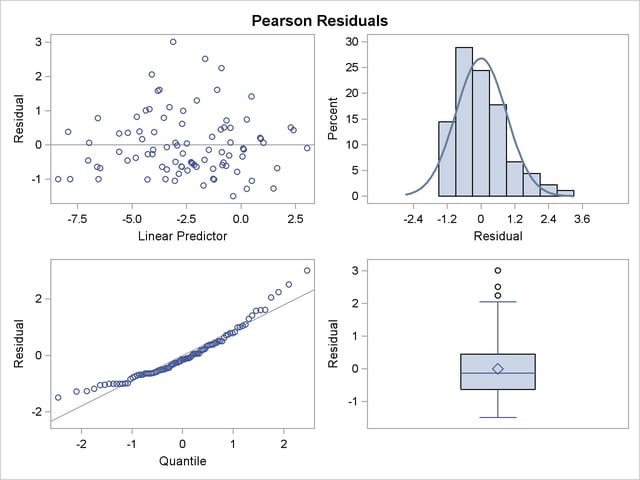
This analysis depends on your choice for the variance function that was implied by the binomial distribution. You can diagnose the distributional assumption by examining various graphical diagnotics measures. The following statements request a panel display of the Pearson-type residuals:
ods graphics on; ods select PearsonPanel; proc glimmix data=blotch plots=pearsonpanel; class site variety; model prop = site variety / link=logit dist=binomial; random _residual_; run; ods graphics off;
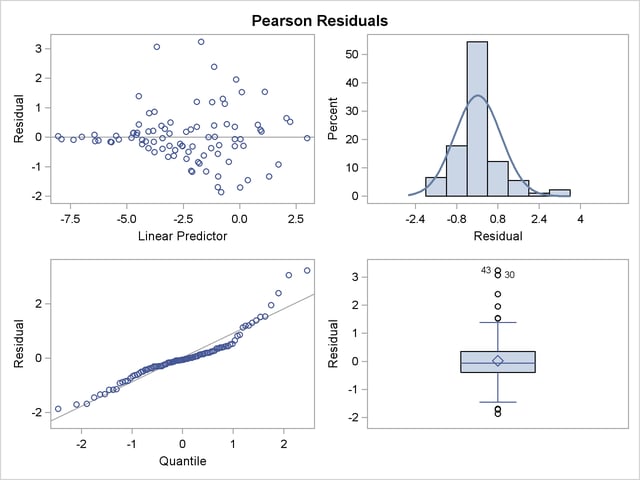
Output 38.4.7 clearly indicates that the chosen variance function is not appropriate for these data. As 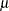 approaches zero or one, the variability in the residuals is less than that implied by the binomial variance function.
approaches zero or one, the variability in the residuals is less than that implied by the binomial variance function.
To remedy this situation, McCullagh and Nelder (1989) consider instead the variance function
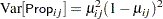 |
Imagine two varieties with 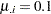 and
and 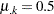 . Under the binomial variance function, the variance of the proportion for variety
. Under the binomial variance function, the variance of the proportion for variety 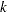 is 2.77 times larger than that for variety
is 2.77 times larger than that for variety 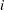 . Under the revised model this ratio increases to
. Under the revised model this ratio increases to 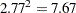 .
.
The analysis of the revised model is obtained with the next set of GLIMMIX statements. Because you need to model a variance function that does not correspond to any of the built-in distributions, you need to supply a function with an assignment to the automatic variable _VARIANCE_. The GLIMMIX procedure then considers the distribution of the data as unknown. The corresponding estimation technique is quasi-likelihood. Because this model does not include an extra scale parameter, you can drop the RANDOM _RESIDUAL_ statement from the analysis.
ods graphics on;
ods select ModelInfo FitStatistics LSMeans Diffs PearsonPanel;
proc glimmix data=blotch plots=pearsonpanel;
class site variety;
_variance_ = _mu_**2 * (1-_mu_)**2;
model prop = site variety / link=logit;
lsmeans variety / diff=control('1');
run;
ods graphics off;
The "Model Information" table in Output 38.4.8 now displays the distribution as "Unknown," because of the assignment made in the GLIMMIX statements to _VARIANCE_. The table also shows the expression evaluated as the variance function.
The fit statistics of the model are now expressed in terms of the log quasi-likelihood. It is computed as
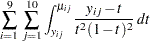 |
Twice the negative of this sum equals 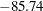 , which is displayed in the "Fit Statistics" table (Output 38.4.9).
, which is displayed in the "Fit Statistics" table (Output 38.4.9).
The scaled Pearson statistic is now 0.99. Inclusion of an extra scale parameter 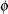 would have little or no effect on the results.
would have little or no effect on the results.
The panel of Pearson-type residuals now shows a much more adequate distribution for the residuals and a reduction in the number of outlying residuals (Output 38.4.10).
The least squares means are no longer ordered in size by variety (Output 38.4.11). For example, 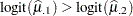 . Under the revised model, the second variety has a greater percentage of its leaf area covered by blotch, compared to the first variety. Varieties 5 and 6 and varieties 8 and 9 show similar reversal in ranking.
. Under the revised model, the second variety has a greater percentage of its leaf area covered by blotch, compared to the first variety. Varieties 5 and 6 and varieties 8 and 9 show similar reversal in ranking.
| variety Least Squares Means | |||||
|---|---|---|---|---|---|
| variety | Estimate | Standard Error | DF | t Value | Pr > |t| |
| 1 | -4.0453 | 0.3333 | 72 | -12.14 | <.0001 |
| 2 | -4.5126 | 0.3333 | 72 | -13.54 | <.0001 |
| 3 | -3.9664 | 0.3333 | 72 | -11.90 | <.0001 |
| 4 | -3.0912 | 0.3333 | 72 | -9.27 | <.0001 |
| 5 | -2.6927 | 0.3333 | 72 | -8.08 | <.0001 |
| 6 | -2.7167 | 0.3333 | 72 | -8.15 | <.0001 |
| 7 | -1.7052 | 0.3333 | 72 | -5.12 | <.0001 |
| 8 | -0.7827 | 0.3333 | 72 | -2.35 | 0.0216 |
| 9 | -0.9098 | 0.3333 | 72 | -2.73 | 0.0080 |
| 10 | -0.1580 | 0.3333 | 72 | -0.47 | 0.6369 |
Interestingly, the standard errors are constant among the LS-means (Output 38.4.11) and among the LS-means differences (Output 38.4.12). This is due to the fact that for the logit link
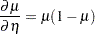 |
which cancels with the square root of the variance function in the estimating equations. The analysis is thus orthogonal.
| Differences of variety Least Squares Means | ||||||
|---|---|---|---|---|---|---|
| variety | _variety | Estimate | Standard Error | DF | t Value | Pr > |t| |
| 2 | 1 | -0.4673 | 0.4714 | 72 | -0.99 | 0.3249 |
| 3 | 1 | 0.07885 | 0.4714 | 72 | 0.17 | 0.8676 |
| 4 | 1 | 0.9541 | 0.4714 | 72 | 2.02 | 0.0467 |
| 5 | 1 | 1.3526 | 0.4714 | 72 | 2.87 | 0.0054 |
| 6 | 1 | 1.3286 | 0.4714 | 72 | 2.82 | 0.0062 |
| 7 | 1 | 2.3401 | 0.4714 | 72 | 4.96 | <.0001 |
| 8 | 1 | 3.2626 | 0.4714 | 72 | 6.92 | <.0001 |
| 9 | 1 | 3.1355 | 0.4714 | 72 | 6.65 | <.0001 |
| 10 | 1 | 3.8873 | 0.4714 | 72 | 8.25 | <.0001 |
Copyright © SAS Institute, Inc. All Rights Reserved.