| The GENMOD Procedure |
Example 37.11 Exact Poisson Regression
The following data, taken from Cox and Snell (1989, pp. 10–11), consists of the number, Notready, of ingots that are not ready for rolling, out of Total tested, for several combinations of heating time and soaking time:
data ingots; input Heat Soak Notready Total @@; lnTotal= log(Total); datalines; 7 1.0 0 10 14 1.0 0 31 27 1.0 1 56 51 1.0 3 13 7 1.7 0 17 14 1.7 0 43 27 1.7 4 44 51 1.7 0 1 7 2.2 0 7 14 2.2 2 33 27 2.2 0 21 51 2.2 0 1 7 2.8 0 12 14 2.8 0 31 27 2.8 1 22 51 4.0 0 1 7 4.0 0 9 14 4.0 0 19 27 4.0 1 16 ;
The following invocation of PROC GENMOD fits an asymptotic (unconditional) Poisson regression model to the data. The variable Notready is specified as the response variable, and the continuous predictors Heat and Soak are defined in the CLASS statement as categorical predictors that use reference coding. Specifying the offset variable as lnTotal enables you to model the ratio Notready/Total.
proc genmod data=ingots; class Heat Soak / param=ref; model Notready=Heat Soak / offset=lnTotal dist=poisson link=log; exact Heat Soak / joint estimate; exactoptions statustime=10; run;
The EXACT statement is specified to additionally fit an exact conditional Poisson regression model. Specifying the lnTotal offset variable models the ratio Notready/Total; in this case, the Total variable contains the largest possible response value for each observation. The JOINT option produces a joint test for the significance of the covariates, along with the usual marginal tests. The ESTIMATE option produces exact parameter estimates for the covariates. The STATUSTIME=10 option is specified in the EXACTOPTIONS statement for monitoring the progress of the results; this example can take several minutes to complete due to the JOINT option. If you run out of memory, see the SAS Companion for your system for information about how to increase the available memory.
The "Criteria For Assessing Goodness Of Fit" table is displayed in Output 37.11.1. Comparing the deviance of 10.9363 with its asymptotic chi-square with 11 degrees of freedom distribution, you find that the 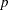 -value is 0.084. This indicates that the specified model fits the data reasonably well.
-value is 0.084. This indicates that the specified model fits the data reasonably well.
| Criteria For Assessing Goodness Of Fit | |||
|---|---|---|---|
| Criterion | DF | Value | Value/DF |
| Deviance | 11 | 10.9363 | 0.9942 |
| Scaled Deviance | 11 | 10.9363 | 0.9942 |
| Pearson Chi-Square | 11 | 9.3722 | 0.8520 |
| Scaled Pearson X2 | 11 | 9.3722 | 0.8520 |
| Log Likelihood | -7.2408 | ||
| Full Log Likelihood | -12.9038 | ||
| AIC (smaller is better) | 41.8076 | ||
| AICC (smaller is better) | 56.2076 | ||
| BIC (smaller is better) | 49.3631 | ||
From the "Analysis Of Parameter Estimates" table in Output 37.11.2, you can see that only two of the Heat parameters are deemed significant. Looking at the standard errors, you can see that the unconditional analysis had convergence difficulties with the Heat=7 parameter (Standard Error=264324.6), which means you cannot fit this unconditional Poisson regression model to this data.
| Analysis Of Maximum Likelihood Parameter Estimates | ||||||||
|---|---|---|---|---|---|---|---|---|
| Parameter | DF | Estimate | Standard Error | Wald 95% Confidence Limits | Wald Chi-Square | Pr > ChiSq | ||
| Intercept | 1 | -1.5700 | 1.1657 | -3.8548 | 0.7147 | 1.81 | 0.1780 | |
| Heat | 7 | 1 | -27.6129 | 264324.6 | -518094 | 518039.0 | 0.00 | 0.9999 |
| Heat | 14 | 1 | -3.0107 | 1.0025 | -4.9756 | -1.0458 | 9.02 | 0.0027 |
| Heat | 27 | 1 | -1.7180 | 0.7691 | -3.2253 | -0.2106 | 4.99 | 0.0255 |
| Soak | 1 | 1 | -0.2454 | 1.1455 | -2.4906 | 1.9998 | 0.05 | 0.8304 |
| Soak | 1.7 | 1 | 0.5572 | 1.1217 | -1.6412 | 2.7557 | 0.25 | 0.6193 |
| Soak | 2.2 | 1 | 0.4079 | 1.2260 | -1.9951 | 2.8109 | 0.11 | 0.7394 |
| Soak | 2.8 | 1 | -0.1301 | 1.4234 | -2.9199 | 2.6597 | 0.01 | 0.9272 |
| Scale | 0 | 1.0000 | 0.0000 | 1.0000 | 1.0000 | |||
| Note: | The scale parameter was held fixed. |
Following the output from the asymptotic analysis, the exact conditional Poisson regression results are displayed, as shown in Output 37.11.3.
| Conditional Exact Tests | ||||
|---|---|---|---|---|
| Effect | Test | Statistic | p-Value | |
| Exact | Mid | |||
| Joint | Score | 18.3665 | 0.0137 | 0.0137 |
| Probability | 1.294E-6 | 0.0471 | 0.0471 | |
| Heat | Score | 15.8259 | 0.0023 | 0.0022 |
| Probability | 0.000175 | 0.0063 | 0.0062 | |
| Soak | Score | 1.4612 | 0.8683 | 0.8646 |
| Probability | 0.00735 | 0.8176 | 0.8139 | |
The Joint test in the "Conditional Exact Tests" table in Output 37.11.3 is produced by specifying the JOINT option in the EXACT statement. The 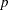 -values for this test indicate that the parameters for Heat and Soak are jointly significant as explanatory effects in the model. If the Heat variable is the only explanatory variable in your model, then the rows of this table labeled as "Heat" show the joint significance of all the Heat effect parameters in that reduced model. In this case, a model that contains only the Heat parameters still explains a significant amount of the variability; however, you can see that a model that contains only the Soak parameters would not be significant.
-values for this test indicate that the parameters for Heat and Soak are jointly significant as explanatory effects in the model. If the Heat variable is the only explanatory variable in your model, then the rows of this table labeled as "Heat" show the joint significance of all the Heat effect parameters in that reduced model. In this case, a model that contains only the Heat parameters still explains a significant amount of the variability; however, you can see that a model that contains only the Soak parameters would not be significant.
The "Exact Parameter Estimates" table in Output 37.11.4 displays parameter estimates and tests of significance for the levels of the CLASS variables. Again, the Heat=7 parameter has some difficulties; however, in the exact analysis, a median unbiased estimate is computed for the parameter instead of a maximum likelihood estimate. The confidence limits show that the Heat variable contains some explanatory power, while the categorical Soak variable is insignificant and can be dropped from the model.
| Exact Parameter Estimates | |||||||
|---|---|---|---|---|---|---|---|
| Parameter | Estimate | Standard Error | 95% Confidence Limits | p-Value | |||
| Heat | 7 | -2.7552 | * | . | -Infinity | -0.4267 | 0.0199 |
| Heat | 14 | -3.0255 | 1.0128 | -5.7450 | -0.6194 | 0.0113 | |
| Heat | 27 | -1.7846 | 0.8065 | -3.6779 | 0.2260 | 0.0844 | |
| Soak | 1 | -0.3231 | 1.1717 | -2.8673 | 3.6754 | 1.0000 | |
| Soak | 1.7 | 0.5375 | 1.1284 | -1.8056 | 4.4588 | 1.0000 | |
| Soak | 2.2 | 0.4035 | 1.2347 | -2.5785 | 4.5054 | 1.0000 | |
| Soak | 2.8 | -0.1661 | 1.4214 | -4.5490 | 4.2168 | 1.0000 | |
| Note: | * indicates a median unbiased estimate. |
Note:If you want to make predictions from the exact results, you can obtain an estimate for the intercept parameter by specifying the INTERCEPT keyword in the EXACT statement. You should also remove the JOINT option to reduce the amount of time and memory consumed.
Copyright © SAS Institute, Inc. All Rights Reserved.