| The GENMOD Procedure |
Example 37.7 Log-Linear Model for Count Data
In this example the data, from Thall and Vail (1990), concern the treatment of people suffering from epileptic seizure episodes. These data are also analyzed in Diggle, Liang, and Zeger (1994). The data consist of the number of epileptic seizures in an eight-week baseline period, before any treatment, and in each of four two-week treatment periods, in which patients received either a placebo or the drug Progabide in addition to other therapy. A portion of the data is displayed in Table 37.12. See "Gee Model for Count Data, Exchangeable Correlation" in the SAS/STAT Sample Program Library for the complete data set.
Patient ID |
Treatment |
Baseline |
Visit1 |
Visit2 |
Visit3 |
Visit4 |
|---|---|---|---|---|---|---|
104 |
Placebo |
11 |
5 |
3 |
3 |
3 |
106 |
Placebo |
11 |
3 |
5 |
3 |
3 |
107 |
Placebo |
6 |
2 |
4 |
0 |
5 |
. |
||||||
. |
||||||
. |
||||||
101 |
Progabide |
76 |
11 |
14 |
9 |
8 |
102 |
Progabide |
38 |
8 |
7 |
9 |
4 |
103 |
Progabide |
19 |
0 |
4 |
3 |
0 |
. |
||||||
. |
||||||
. |
Model the data as a log-linear model with 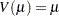 (the Poisson variance function) and
(the Poisson variance function) and
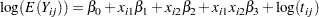 |
where
 number of epileptic seizures in interval
number of epileptic seizures in interval 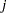
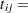 length of interval
length of interval 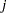
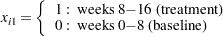

The correlations between the counts are modeled as 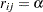 ,
,  (exchangeable correlations). For comparison, the correlations are also modeled as independent (identity correlation matrix). In this model, the regression parameters have the interpretation in terms of the log seizure rate displayed in Table 37.13.
(exchangeable correlations). For comparison, the correlations are also modeled as independent (identity correlation matrix). In this model, the regression parameters have the interpretation in terms of the log seizure rate displayed in Table 37.13.
Treatment |
Visit |
|
|---|---|---|
Placebo |
Baseline |
|
1–4 |
|
|
Progabide |
Baseline |
|
1–4 |
|
The difference between the log seizure rates in the pretreatment (baseline) period and the treatment periods is 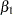 for the placebo group and
for the placebo group and 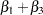 for the Progabide group. A value of
for the Progabide group. A value of  indicates a reduction in the seizure rate.
indicates a reduction in the seizure rate.
Output 37.7.1 lists the first 14 observations of the data, which are arranged as one visit per observation:
Some further data manipulations create an observation for the baseline measures, a log time interval variable for use as an offset, and an indicator variable for whether the observation is for a baseline measurement or a visit measurement. Patient 207 is deleted as an outlier, as in the Diggle, Liang, and Zeger (1994) analysis.
The following statements prepare the data for analysis with PROC GENMOD:
data new;
set thall;
output;
if visit=1 then do;
y=bline;
visit=0;
output;
end;
run;
data new;
set new;
if id ne 207;
if visit=0 then do;
x1=0;
ltime=log(8);
end;
else do;
x1=1;
ltime=log(2);
end;
run;
For comparison with the GEE results, an ordinary Poisson regression is first fit. The results are shown in Output 37.7.2.
| Analysis Of Maximum Likelihood Parameter Estimates | |||||||
|---|---|---|---|---|---|---|---|
| Parameter | DF | Estimate | Standard Error | Wald 95% Confidence Limits | Wald Chi-Square | Pr > ChiSq | |
| Intercept | 1 | 1.3476 | 0.0341 | 1.2809 | 1.4144 | 1565.44 | <.0001 |
| x1 | 1 | 0.1108 | 0.0469 | 0.0189 | 0.2027 | 5.58 | 0.0181 |
| trt | 1 | -0.1080 | 0.0486 | -0.2034 | -0.0127 | 4.93 | 0.0264 |
| x1*trt | 1 | -0.3016 | 0.0697 | -0.4383 | -0.1649 | 18.70 | <.0001 |
| Scale | 0 | 1.0000 | 0.0000 | 1.0000 | 1.0000 | ||
| Note: | The scale parameter was held fixed. |
The GEE solution is requested with the REPEATED statement in the GENMOD procedure. The SUBJECT=ID option indicates that the variable id describes the observations for a single cluster, and the CORRW option displays the working correlation matrix. The TYPE= option specifies the correlation structure; the value EXCH indicates the exchangeable structure.
The following statements perform the analysis:
proc genmod data=new; class id; model y=x1 | trt / d=poisson offset=ltime; repeated subject=id / corrw covb type=exch; run;
These statements first fit a generalized linear model (GLM) to these data by maximum likelihood. The estimates are not shown in the output, but are used as initial values for the GEE solution.
Information about the GEE model is displayed in Output 37.7.3. The results of fitting the model are displayed in Output 37.7.4. Compare these with the model of independence displayed in Output 37.7.2. The parameter estimates are nearly identical, but the standard errors for the independence case are underestimated. The coefficient of the interaction term,  , is highly significant under the independence model and marginally significant with the exchangeable correlations model.
, is highly significant under the independence model and marginally significant with the exchangeable correlations model.
| Analysis Of GEE Parameter Estimates | ||||||
|---|---|---|---|---|---|---|
| Empirical Standard Error Estimates | ||||||
| Parameter | Estimate | Standard Error | 95% Confidence Limits | Z | Pr > |Z| | |
| Intercept | 1.3476 | 0.1574 | 1.0392 | 1.6560 | 8.56 | <.0001 |
| x1 | 0.1108 | 0.1161 | -0.1168 | 0.3383 | 0.95 | 0.3399 |
| trt | -0.1080 | 0.1937 | -0.4876 | 0.2716 | -0.56 | 0.5770 |
| x1*trt | -0.3016 | 0.1712 | -0.6371 | 0.0339 | -1.76 | 0.0781 |
Table 37.14 displays the regression coefficients, standard errors, and normalized coefficients that result from fitting the model with independent and exchangeable working correlation matrices.
Variable |
Correlation Structure |
Coef. |
Std. Error |
Coef./S.E. |
|---|---|---|---|---|
Intercept |
Exchangeable |
1.35 |
0.16 |
8.56 |
Independent |
1.35 |
0.03 |
39.52 |
|
Visit |
Exchangeable |
0.11 |
0.12 |
0.95 |
Independent |
0.11 |
0.05 |
2.36 |
|
Treat |
Exchangeable |
|
0.19 |
|
Independent |
|
0.05 |
|
|
|
Exchangeable |
|
0.17 |
|
Independent |
|
0.07 |
|
The fitted exchangeable correlation matrix is specified with the CORRW option and is displayed in Output 37.7.5.
If you specify the COVB option, you produce both the model-based (naive) and the empirical (robust) covariance matrices. Output 37.7.6 contains these estimates.
The two covariance estimates are similar, indicating an adequate correlation model.
Copyright © SAS Institute, Inc. All Rights Reserved.
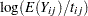

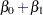
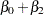
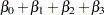
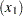
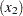
 0.11
0.11 