| The CLUSTER Procedure |
Example 29.2 Crude Birth and Death Rates
This example uses the SAS data set Poverty created in the section Getting Started: CLUSTER Procedure. The data, from Rouncefield (1995), are birth rates, death rates, and infant death rates for 97 countries. Six cluster analyses are performed with eight methods. Scatter plots showing cluster membership at selected levels are produced instead of tree diagrams.
Each cluster analysis is performed by a macro called ANALYZE. The macro takes two arguments. The first, &METHOD, specifies the value of the METHOD= option to be used in the PROC CLUSTER statement. The second, &NCL, must be specified as a list of integers, separated by blanks, indicating the number of clusters desired in each scatter plot. For example, the first invocation of ANALYZE specifies the AVERAGE method and requests plots of 3 and 8 clusters. When two-stage density linkage is used, the K= and R= options are specified as part of the first argument.
The ANALYZE macro first invokes the CLUSTER procedure with METHOD=&METHOD, where &METHOD represents the value of the first argument to ANALYZE. This part of the macro produces the PROC CLUSTER output shown.
The %DO loop processes &NCL, the list of numbers of clusters to plot. The macro variable &K is a counter that indexes the numbers within &NCL. The %SCAN function picks out the &Kth number in &NCL, which is then assigned to the macro variable &N. When &K exceeds the number of numbers in &NCL, %SCAN returns a null string. Thus, the %DO loop executes while &N is not equal to a null string. In the %WHILE condition, a null string is indicated by the absence of any nonblank characters between the comparison operator (NE) and the right parenthesis that terminates the condition.
Within the %DO loop, the TREE procedure creates an output data set containing &N clusters. The SGPLOT procedure then produces a scatter plot in which each observation is identified by the number of the cluster to which it belongs. The TITLE2 statement uses double quotes so that &N and &METHOD can be used within the title. At the end of the loop, &K is incremented by 1, and the next number is extracted from &NCL by %SCAN.
title 'Cluster Analysis of Birth and Death Rates';
ods graphics on;
%macro analyze(method,ncl);
proc cluster data=poverty outtree=tree method=&method print=15 ccc pseudo;
var birth death;
title2;
run;
%let k=1;
%let n=%scan(&ncl,&k);
%do %while(&n NE);
proc tree data=tree noprint out=out ncl=&n;
copy birth death;
run;
proc sgplot;
scatter y=death x=birth / group=cluster;
title2 "Plot of &n Clusters from METHOD=&METHOD";
run;
%let k=%eval(&k+1);
%let n=%scan(&ncl,&k);
%end;
%mend;
The following statement produces Output 29.2.1, Output 29.2.3, and Output 29.2.4:
%analyze(average, 3 8)
For average linkage, the CCC has peaks at 3, 8, 10, and 12 clusters, but the 3-cluster peak is lower than the 8-cluster peak. The pseudo 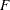 statistic has peaks at 3, 8, and 12 clusters. The pseudo
statistic has peaks at 3, 8, and 12 clusters. The pseudo 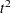 statistic drops sharply at 3 clusters, continues to fall at 4 clusters, and has a particularly low value at 12 clusters. However, there are not enough data to seriously consider as many as 12 clusters. Scatter plots are given for 3 and 8 clusters. The results are shown in Output 29.2.1 through Output 29.2.4. In Output 29.2.4, the eighth cluster consists of the two outlying observations, Mexico and Korea.
statistic drops sharply at 3 clusters, continues to fall at 4 clusters, and has a particularly low value at 12 clusters. However, there are not enough data to seriously consider as many as 12 clusters. Scatter plots are given for 3 and 8 clusters. The results are shown in Output 29.2.1 through Output 29.2.4. In Output 29.2.4, the eighth cluster consists of the two outlying observations, Mexico and Korea.
| Cluster Analysis of Birth and Death Rates |
| Eigenvalues of the Covariance Matrix | ||||
|---|---|---|---|---|
| Eigenvalue | Difference | Proportion | Cumulative | |
| 1 | 189.106588 | 173.101020 | 0.9220 | 0.9220 |
| 2 | 16.005568 | 0.0780 | 1.0000 | |
| Cluster History | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| NCL | Clusters Joined | FREQ | SPRSQ | RSQ | ERSQ | CCC | PSF | PST2 | Norm RMS Dist |
T i e |
|
| 15 | CL27 | CL20 | 18 | 0.0035 | .980 | .975 | 2.61 | 292 | 18.6 | 0.2325 | |
| 14 | CL23 | CL17 | 28 | 0.0034 | .977 | .972 | 1.97 | 271 | 17.7 | 0.2358 | |
| 13 | CL18 | CL54 | 8 | 0.0015 | .975 | .969 | 2.35 | 279 | 7.1 | 0.2432 | |
| 12 | CL21 | CL26 | 8 | 0.0015 | .974 | .966 | 2.85 | 290 | 6.1 | 0.2493 | |
| 11 | CL19 | CL24 | 12 | 0.0033 | .971 | .962 | 2.78 | 285 | 14.8 | 0.2767 | |
| 10 | CL22 | CL16 | 12 | 0.0036 | .967 | .957 | 2.84 | 284 | 17.4 | 0.2858 | |
| 9 | CL15 | CL28 | 22 | 0.0061 | .961 | .951 | 2.45 | 271 | 17.5 | 0.3353 | |
| 8 | OB23 | OB61 | 2 | 0.0014 | .960 | .943 | 3.59 | 302 | . | 0.3703 | |
| 7 | CL25 | CL11 | 17 | 0.0098 | .950 | .933 | 3.01 | 284 | 23.3 | 0.4033 | |
| 6 | CL7 | CL12 | 25 | 0.0122 | .938 | .920 | 2.63 | 273 | 14.8 | 0.4132 | |
| 5 | CL10 | CL14 | 40 | 0.0303 | .907 | .902 | 0.59 | 225 | 82.7 | 0.4584 | |
| 4 | CL13 | CL6 | 33 | 0.0244 | .883 | .875 | 0.77 | 234 | 22.2 | 0.5194 | |
| 3 | CL9 | CL8 | 24 | 0.0182 | .865 | .827 | 2.13 | 300 | 27.7 | 0.735 | |
| 2 | CL5 | CL3 | 64 | 0.1836 | .681 | .697 | -.55 | 203 | 148 | 0.8402 | |
| 1 | CL2 | CL4 | 97 | 0.6810 | .000 | .000 | 0.00 | . | 203 | 1.3348 | |



The following statement produces Output 29.2.5 and Output 29.2.7:
%analyze(complete, 3)
Complete linkage shows CCC peaks at 3, 8 and 12 clusters. The pseudo 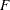 statistic peaks at 3 and 12 clusters. The pseudo
statistic peaks at 3 and 12 clusters. The pseudo 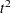 statistic indicates 3 clusters.
statistic indicates 3 clusters.
The scatter plot for 3 clusters is shown.
| Cluster Analysis of Birth and Death Rates |
| Eigenvalues of the Covariance Matrix | ||||
|---|---|---|---|---|
| Eigenvalue | Difference | Proportion | Cumulative | |
| 1 | 189.106588 | 173.101020 | 0.9220 | 0.9220 |
| 2 | 16.005568 | 0.0780 | 1.0000 | |
| Cluster History | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| NCL | Clusters Joined | FREQ | SPRSQ | RSQ | ERSQ | CCC | PSF | PST2 | Norm Max Dist |
T i e |
|
| 15 | CL22 | CL33 | 8 | 0.0015 | .983 | .975 | 3.80 | 329 | 6.1 | 0.4092 | |
| 14 | CL56 | CL18 | 8 | 0.0014 | .981 | .972 | 3.97 | 331 | 6.6 | 0.4255 | |
| 13 | CL30 | CL44 | 8 | 0.0019 | .979 | .969 | 4.04 | 330 | 19.0 | 0.4332 | |
| 12 | OB23 | OB61 | 2 | 0.0014 | .978 | .966 | 4.45 | 340 | . | 0.4378 | |
| 11 | CL19 | CL24 | 24 | 0.0034 | .974 | .962 | 4.17 | 327 | 24.1 | 0.4962 | |
| 10 | CL17 | CL28 | 12 | 0.0033 | .971 | .957 | 4.18 | 325 | 14.8 | 0.5204 | |
| 9 | CL20 | CL13 | 16 | 0.0067 | .964 | .951 | 3.38 | 297 | 25.2 | 0.5236 | |
| 8 | CL11 | CL21 | 32 | 0.0054 | .959 | .943 | 3.44 | 297 | 19.7 | 0.6001 | |
| 7 | CL26 | CL15 | 13 | 0.0096 | .949 | .933 | 2.93 | 282 | 28.9 | 0.7233 | |
| 6 | CL14 | CL10 | 20 | 0.0128 | .937 | .920 | 2.46 | 269 | 27.7 | 0.8033 | |
| 5 | CL9 | CL16 | 30 | 0.0237 | .913 | .902 | 1.29 | 241 | 47.1 | 0.8993 | |
| 4 | CL6 | CL7 | 33 | 0.0240 | .889 | .875 | 1.38 | 248 | 21.7 | 1.2165 | |
| 3 | CL5 | CL12 | 32 | 0.0178 | .871 | .827 | 2.56 | 317 | 13.6 | 1.2326 | |
| 2 | CL3 | CL8 | 64 | 0.1900 | .681 | .697 | -.55 | 203 | 167 | 1.5412 | |
| 1 | CL2 | CL4 | 97 | 0.6810 | .000 | .000 | 0.00 | . | 203 | 2.5233 | |


The following statement produces Output 29.2.8 and Output 29.2.10:
%analyze(single, 7 10)
The CCC and pseudo 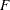 statistics are not appropriate for use with single linkage because of the method’s tendency to chop off tails of distributions. The pseudo
statistics are not appropriate for use with single linkage because of the method’s tendency to chop off tails of distributions. The pseudo 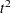 statistic can be used by looking for large values and taking the number of clusters to be one greater than the level at which the large pseudo
statistic can be used by looking for large values and taking the number of clusters to be one greater than the level at which the large pseudo 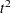 value is displayed. For these data, there are large values at levels 6 and 9, suggesting 7 or 10 clusters.
value is displayed. For these data, there are large values at levels 6 and 9, suggesting 7 or 10 clusters.
The scatter plots for 7 and 10 clusters are shown.
| Cluster Analysis of Birth and Death Rates |
| Eigenvalues of the Covariance Matrix | ||||
|---|---|---|---|---|
| Eigenvalue | Difference | Proportion | Cumulative | |
| 1 | 189.106588 | 173.101020 | 0.9220 | 0.9220 |
| 2 | 16.005568 | 0.0780 | 1.0000 | |
| Cluster History | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| NCL | Clusters Joined | FREQ | SPRSQ | RSQ | ERSQ | CCC | PSF | PST2 | Norm Min Dist |
T i e |
|
| 15 | CL37 | CL19 | 8 | 0.0014 | .968 | .975 | -2.3 | 178 | 6.6 | 0.1331 | |
| 14 | CL20 | CL23 | 15 | 0.0059 | .962 | .972 | -3.1 | 162 | 18.7 | 0.1412 | |
| 13 | CL14 | CL16 | 19 | 0.0054 | .957 | .969 | -3.4 | 155 | 8.8 | 0.1442 | |
| 12 | CL26 | OB58 | 31 | 0.0014 | .955 | .966 | -2.7 | 165 | 4.0 | 0.1486 | |
| 11 | OB86 | CL18 | 4 | 0.0003 | .955 | .962 | -1.6 | 183 | 3.8 | 0.1495 | |
| 10 | CL13 | CL11 | 23 | 0.0088 | .946 | .957 | -2.3 | 170 | 11.3 | 0.1518 | |
| 9 | CL22 | CL17 | 30 | 0.0235 | .923 | .951 | -4.7 | 131 | 45.7 | 0.1593 | T |
| 8 | CL15 | CL10 | 31 | 0.0210 | .902 | .943 | -5.8 | 117 | 21.8 | 0.1593 | |
| 7 | CL9 | OB75 | 31 | 0.0052 | .897 | .933 | -4.7 | 130 | 4.0 | 0.1628 | |
| 6 | CL7 | CL12 | 62 | 0.2023 | .694 | .920 | -15 | 41.3 | 223 | 0.1725 | |
| 5 | CL6 | CL8 | 93 | 0.6681 | .026 | .902 | -26 | 0.6 | 199 | 0.1756 | |
| 4 | CL5 | OB48 | 94 | 0.0056 | .021 | .875 | -24 | 0.7 | 0.5 | 0.1811 | T |
| 3 | CL4 | OB67 | 95 | 0.0083 | .012 | .827 | -15 | 0.6 | 0.8 | 0.1811 | |
| 2 | OB23 | OB61 | 2 | 0.0014 | .011 | .697 | -13 | 1.0 | . | 0.4378 | |
| 1 | CL3 | CL2 | 97 | 0.0109 | .000 | .000 | 0.00 | . | 1.0 | 0.5815 | |



The following statements produce Output 29.2.11 through Output 29.2.14, :
%analyze(two k=10, 3)
%analyze(two k=18, 2)
For 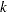 th-nearest-neighbor density linkage, the number of modes as a function of
th-nearest-neighbor density linkage, the number of modes as a function of 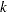 is as follows (not all of these analyses are shown):
is as follows (not all of these analyses are shown):
|
modes |
|
3 |
13 |
|
4 |
6 |
|
5-7 |
4 |
|
8-15 |
3 |
|
16-21 |
2 |
|
22+ |
1 |
Thus, there is strong evidence of 3 modes and an indication of the possibility of 2 modes. Uniform-kernel density linkage gives similar results. For K=10 (10th-nearest-neighbor density linkage), the scatter plot for 3 clusters is shown; and for K=18, the scatter plot for 2 clusters is shown.
| Cluster Analysis of Birth and Death Rates |
| Eigenvalues of the Covariance Matrix | ||||
|---|---|---|---|---|
| Eigenvalue | Difference | Proportion | Cumulative | |
| 1 | 189.106588 | 173.101020 | 0.9220 | 0.9220 |
| 2 | 16.005568 | 0.0780 | 1.0000 | |
| Cluster History | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCL | FREQ | SPRSQ | RSQ | ERSQ | CCC | PSF | PST2 | Normalized Fusion Density |
Maximum Density in Each Cluster |
T i e |
|||
| Clusters Joined | Lesser | Greater | |||||||||||
| 15 | CL16 | OB94 | 22 | 0.0015 | .921 | .975 | -11 | 68.4 | 1.4 | 9.2234 | 6.7927 | 15.3069 | |
| 14 | CL19 | OB49 | 28 | 0.0021 | .919 | .972 | -11 | 72.4 | 1.8 | 8.7369 | 5.9334 | 33.4385 | |
| 13 | CL15 | OB52 | 23 | 0.0024 | .917 | .969 | -10 | 76.9 | 2.3 | 8.5847 | 5.9651 | 15.3069 | |
| 12 | CL13 | OB96 | 24 | 0.0018 | .915 | .966 | -9.3 | 83.0 | 1.6 | 7.9252 | 5.4724 | 15.3069 | |
| 11 | CL12 | OB93 | 25 | 0.0025 | .912 | .962 | -8.5 | 89.5 | 2.2 | 7.8913 | 5.4401 | 15.3069 | |
| 10 | CL11 | OB78 | 26 | 0.0031 | .909 | .957 | -7.7 | 96.9 | 2.5 | 7.787 | 5.4082 | 15.3069 | |
| 9 | CL10 | OB76 | 27 | 0.0026 | .907 | .951 | -6.7 | 107 | 2.1 | 7.7133 | 5.4401 | 15.3069 | |
| 8 | CL9 | OB77 | 28 | 0.0023 | .904 | .943 | -5.5 | 120 | 1.7 | 7.4256 | 4.9017 | 15.3069 | |
| 7 | CL8 | OB43 | 29 | 0.0022 | .902 | .933 | -4.1 | 138 | 1.6 | 6.927 | 4.4764 | 15.3069 | |
| 6 | CL7 | OB87 | 30 | 0.0043 | .898 | .920 | -2.7 | 160 | 3.1 | 4.932 | 2.9977 | 15.3069 | |
| 5 | CL6 | OB82 | 31 | 0.0055 | .892 | .902 | -1.1 | 191 | 3.7 | 3.7331 | 2.1560 | 15.3069 | |
| 4 | CL22 | OB61 | 37 | 0.0079 | .884 | .875 | 0.93 | 237 | 10.6 | 3.1713 | 1.6308 | 100.0 | |
| 3 | CL14 | OB23 | 29 | 0.0126 | .872 | .827 | 2.60 | 320 | 10.4 | 2.0654 | 1.0744 | 33.4385 | |
| 2 | CL4 | CL3 | 66 | 0.2129 | .659 | .697 | -1.3 | 183 | 172 | 12.409 | 33.4385 | 100.0 | |
| 1 | CL2 | CL5 | 97 | 0.6588 | .000 | .000 | 0.00 | . | 183 | 10.071 | 15.3069 | 100.0 | |
| Cluster Analysis of Birth and Death Rates |
| Eigenvalues of the Covariance Matrix | ||||
|---|---|---|---|---|
| Eigenvalue | Difference | Proportion | Cumulative | |
| 1 | 189.106588 | 173.101020 | 0.9220 | 0.9220 |
| 2 | 16.005568 | 0.0780 | 1.0000 | |
| Cluster History | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NCL | FREQ | SPRSQ | RSQ | ERSQ | CCC | PSF | PST2 | Normalized Fusion Density |
Maximum Density in Each Cluster |
T i e |
|||
| Clusters Joined | Lesser | Greater | |||||||||||
| 15 | CL16 | OB72 | 46 | 0.0107 | .799 | .975 | -21 | 23.3 | 3.0 | 10.118 | 7.7445 | 23.4457 | |
| 14 | CL15 | OB94 | 47 | 0.0098 | .789 | .972 | -21 | 23.9 | 2.7 | 9.676 | 7.1257 | 23.4457 | |
| 13 | CL14 | OB51 | 48 | 0.0037 | .786 | .969 | -20 | 25.6 | 1.0 | 9.409 | 6.8398 | 23.4457 | T |
| 12 | CL13 | OB96 | 49 | 0.0099 | .776 | .966 | -19 | 26.7 | 2.6 | 9.409 | 6.8398 | 23.4457 | |
| 11 | CL12 | OB76 | 50 | 0.0114 | .764 | .962 | -19 | 27.9 | 2.9 | 8.8136 | 6.3138 | 23.4457 | |
| 10 | CL11 | OB77 | 51 | 0.0021 | .762 | .957 | -18 | 31.0 | 0.5 | 8.6593 | 6.0751 | 23.4457 | |
| 9 | CL10 | OB78 | 52 | 0.0103 | .752 | .951 | -17 | 33.3 | 2.5 | 8.6007 | 6.0976 | 23.4457 | |
| 8 | CL9 | OB43 | 53 | 0.0034 | .748 | .943 | -16 | 37.8 | 0.8 | 8.4964 | 5.9160 | 23.4457 | |
| 7 | CL8 | OB93 | 54 | 0.0109 | .737 | .933 | -15 | 42.1 | 2.6 | 8.367 | 5.7913 | 23.4457 | |
| 6 | CL7 | OB88 | 55 | 0.0110 | .726 | .920 | -13 | 48.3 | 2.6 | 7.916 | 5.3679 | 23.4457 | |
| 5 | CL6 | OB87 | 56 | 0.0120 | .714 | .902 | -12 | 57.5 | 2.7 | 6.6917 | 4.3415 | 23.4457 | |
| 4 | CL20 | OB61 | 39 | 0.0077 | .707 | .875 | -9.8 | 74.7 | 8.3 | 6.2578 | 3.2882 | 100.0 | |
| 3 | CL5 | OB82 | 57 | 0.0138 | .693 | .827 | -5.0 | 106 | 3.0 | 5.3605 | 3.2834 | 23.4457 | |
| 2 | CL3 | OB23 | 58 | 0.0117 | .681 | .697 | -.54 | 203 | 2.5 | 3.2687 | 1.7568 | 23.4457 | |
| 1 | CL2 | CL4 | 97 | 0.6812 | .000 | .000 | 0.00 | . | 203 | 13.764 | 23.4457 | 100.0 | |


In summary, most of the clustering methods indicate 3 or 8 clusters. Most methods agree at the 3-cluster level, but at the other levels, there is considerable disagreement about the composition of the clusters. The presence of numerous ties also complicates the analysis; see Example 29.4.
Copyright © SAS Institute, Inc. All Rights Reserved.