| The MIXED Procedure |
Example 56.4 Known G and R
This animal breeding example from Henderson (1984, p. 48) considers multiple traits. The data are artificial and consist of measurements of two traits on three animals, but the second trait of the third animal is missing. Assuming an additive genetic model, you can use PROC MIXED to predict the breeding value of both traits on all three animals and also to predict the second trait of the third animal. The data are as follows:
data h;
input Trait Animal Y;
datalines;
1 1 6
1 2 8
1 3 7
2 1 9
2 2 5
2 3 .
;
Both 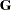 and
and 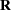 are known.
are known.
 |
 |
In order to read 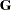 into PROC MIXED by using the GDATA= option in the RANDOM statement, perform the following DATA step:
into PROC MIXED by using the GDATA= option in the RANDOM statement, perform the following DATA step:
data g;
input Row Col1-Col6;
datalines;
1 2 1 1 2 1 1
2 1 2 .5 1 2 .5
3 1 .5 2 1 .5 2
4 2 1 1 3 1.5 1.5
5 1 2 .5 1.5 3 .75
6 1 .5 2 1.5 .75 3
;
The preceding data are in the dense representation for a GDATA= data set. You can also construct a data set with the sparse representation by using Row, Col, and Value variables, although this would require 21 observations instead of 6 for this example.
The PROC MIXED statements are as follows:
proc mixed data=h mmeq mmeqsol;
class Trait Animal;
model Y = Trait / noint s outp=predicted;
random Trait*Animal / type=un gdata=g g gi s;
repeated / type=un sub=Animal r ri;
parms (4) (1) (5) / noiter;
run;
proc print data=predicted;
run;
The MMEQ and MMEQSOL options request the mixed model equations and their solution. The variables Trait and Animal are classification variables, and Trait defines the entire 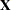 matrix for the fixed-effects portion of the model, since the intercept is omitted with the NOINT option. The fixed-effects solution vector and predicted values are also requested by using the S and OUTP= options, respectively.
matrix for the fixed-effects portion of the model, since the intercept is omitted with the NOINT option. The fixed-effects solution vector and predicted values are also requested by using the S and OUTP= options, respectively.
The random effect Trait*Animal leads to a 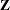 matrix with six columns, the first five corresponding to the identity matrix and the last consisting of 0s. An unstructured
matrix with six columns, the first five corresponding to the identity matrix and the last consisting of 0s. An unstructured 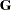 matrix is specified by using the TYPE=UN option, and it is read into PROC MIXED from a SAS data set by using the GDATA=G specification. The G and GI options request the display of
matrix is specified by using the TYPE=UN option, and it is read into PROC MIXED from a SAS data set by using the GDATA=G specification. The G and GI options request the display of 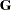 and
and 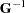 , respectively. The S option requests that the random-effects solution vector be displayed.
, respectively. The S option requests that the random-effects solution vector be displayed.
Note that the preceding 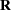 matrix is block diagonal if the data are sorted by animals. The REPEATED statement exploits this fact by requesting
matrix is block diagonal if the data are sorted by animals. The REPEATED statement exploits this fact by requesting 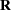 to have unstructured 2
to have unstructured 2 2 blocks corresponding to animals, which are the subjects. The R and RI options request that the estimated 2
2 blocks corresponding to animals, which are the subjects. The R and RI options request that the estimated 2 2 blocks for the first animal and its inverse be displayed. The PARMS statement lists the parameters of this 2
2 blocks for the first animal and its inverse be displayed. The PARMS statement lists the parameters of this 2 2 matrix. Note that the parameters from
2 matrix. Note that the parameters from 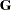 are not specified in the PARMS statement because they have already been assigned by using the GDATA= option in the RANDOM statement. The NOITER option prevents PROC MIXED from computing residual (restricted) maximum likelihood estimates; instead, the known values are used for inferences.
are not specified in the PARMS statement because they have already been assigned by using the GDATA= option in the RANDOM statement. The NOITER option prevents PROC MIXED from computing residual (restricted) maximum likelihood estimates; instead, the known values are used for inferences.
The results from this analysis are shown in Output 56.4.1–Output 56.4.12.
The "Unstructured" covariance structure (Output 56.4.1) applies to both 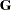 and
and 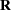 here. The levels of Trait and Animal have been specified correctly.
here. The levels of Trait and Animal have been specified correctly.
| Model Information | |
|---|---|
| Data Set | WORK.H |
| Dependent Variable | Y |
| Covariance Structure | Unstructured |
| Subject Effect | Animal |
| Estimation Method | REML |
| Residual Variance Method | None |
| Fixed Effects SE Method | Model-Based |
| Degrees of Freedom Method | Containment |
The three covariance parameters indicated in Output 56.4.2 correspond to those from the 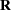 matrix. Those from
matrix. Those from 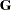 are considered fixed and known because of the GDATA= option.
are considered fixed and known because of the GDATA= option.
Because starting values for the covariance parameters are specified in the PARMS statement, the MIXED procedure prints the residual (restricted) log likelihood at the starting values. Because of the NOITER option in the PARMS statement, this is also the final log likelihood in this analysis (Output 56.4.3).
The block of 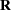 corresponding to the first animal and the inverse of this block are shown in Output 56.4.4.
corresponding to the first animal and the inverse of this block are shown in Output 56.4.4.
The 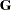 matrix as specified in the GDATA= data set and its inverse are shown in Output 56.4.5 and Output 56.4.6.
matrix as specified in the GDATA= data set and its inverse are shown in Output 56.4.5 and Output 56.4.6.
| Estimated G Matrix | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Row | Effect | Trait | Animal | Col1 | Col2 | Col3 | Col4 | Col5 | Col6 |
| 1 | Trait*Animal | 1 | 1 | 2.0000 | 1.0000 | 1.0000 | 2.0000 | 1.0000 | 1.0000 |
| 2 | Trait*Animal | 1 | 2 | 1.0000 | 2.0000 | 0.5000 | 1.0000 | 2.0000 | 0.5000 |
| 3 | Trait*Animal | 1 | 3 | 1.0000 | 0.5000 | 2.0000 | 1.0000 | 0.5000 | 2.0000 |
| 4 | Trait*Animal | 2 | 1 | 2.0000 | 1.0000 | 1.0000 | 3.0000 | 1.5000 | 1.5000 |
| 5 | Trait*Animal | 2 | 2 | 1.0000 | 2.0000 | 0.5000 | 1.5000 | 3.0000 | 0.7500 |
| 6 | Trait*Animal | 2 | 3 | 1.0000 | 0.5000 | 2.0000 | 1.5000 | 0.7500 | 3.0000 |
| Estimated Inv(G) Matrix | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Row | Effect | Trait | Animal | Col1 | Col2 | Col3 | Col4 | Col5 | Col6 |
| 1 | Trait*Animal | 1 | 1 | 2.5000 | -1.0000 | -1.0000 | -1.6667 | 0.6667 | 0.6667 |
| 2 | Trait*Animal | 1 | 2 | -1.0000 | 2.0000 | 0.6667 | -1.3333 | ||
| 3 | Trait*Animal | 1 | 3 | -1.0000 | 2.0000 | 0.6667 | -1.3333 | ||
| 4 | Trait*Animal | 2 | 1 | -1.6667 | 0.6667 | 0.6667 | 1.6667 | -0.6667 | -0.6667 |
| 5 | Trait*Animal | 2 | 2 | 0.6667 | -1.3333 | -0.6667 | 1.3333 | ||
| 6 | Trait*Animal | 2 | 3 | 0.6667 | -1.3333 | -0.6667 | 1.3333 | ||
The table of covariance parameter estimates in Output 56.4.7 displays only the parameters in 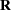 . Because of the GDATA= option in the RANDOM statement, the G-side parameters do not participate in the parameter estimation process. Because of the NOITER option in the PARMS statement, however, the R-side parameters in this output are identical to their starting values.
. Because of the GDATA= option in the RANDOM statement, the G-side parameters do not participate in the parameter estimation process. Because of the NOITER option in the PARMS statement, however, the R-side parameters in this output are identical to their starting values.
The coefficients of the mixed model equations in Output 56.4.8 agree with Henderson (1984, p. 55). Recall from Output 56.4.1 that there are 2 columns in 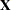 and 6 columns in
and 6 columns in 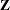 . The first 8 columns of the mixed model equations correspond to the
. The first 8 columns of the mixed model equations correspond to the 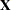 and
and 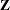 components. Column 9 represents the Y border.
components. Column 9 represents the Y border.
| Mixed Model Equations | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Row | Effect | Trait | Animal | Col1 | Col2 | Col3 | Col4 | Col5 | Col6 | Col7 | Col8 | Col9 |
| 1 | Trait | 1 | 0.7763 | -0.1053 | 0.2632 | 0.2632 | 0.2500 | -0.05263 | -0.05263 | 4.6974 | ||
| 2 | Trait | 2 | -0.1053 | 0.4211 | -0.05263 | -0.05263 | 0.2105 | 0.2105 | 2.2105 | |||
| 3 | Trait*Animal | 1 | 1 | 0.2632 | -0.05263 | 2.7632 | -1.0000 | -1.0000 | -1.7193 | 0.6667 | 0.6667 | 1.1053 |
| 4 | Trait*Animal | 1 | 2 | 0.2632 | -0.05263 | -1.0000 | 2.2632 | 0.6667 | -1.3860 | 1.8421 | ||
| 5 | Trait*Animal | 1 | 3 | 0.2500 | -1.0000 | 2.2500 | 0.6667 | -1.3333 | 1.7500 | |||
| 6 | Trait*Animal | 2 | 1 | -0.05263 | 0.2105 | -1.7193 | 0.6667 | 0.6667 | 1.8772 | -0.6667 | -0.6667 | 1.5789 |
| 7 | Trait*Animal | 2 | 2 | -0.05263 | 0.2105 | 0.6667 | -1.3860 | -0.6667 | 1.5439 | 0.6316 | ||
| 8 | Trait*Animal | 2 | 3 | 0.6667 | -1.3333 | -0.6667 | 1.3333 | |||||
The solution to the mixed model equations also matches that given by Henderson (1984, p. 55). After solving the augmented mixed model equations, you can find the solutions for fixed and random effects in the last column (Output 56.4.9).
| Mixed Model Equations Solution | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Row | Effect | Trait | Animal | Col1 | Col2 | Col3 | Col4 | Col5 | Col6 | Col7 | Col8 | Col9 |
| 1 | Trait | 1 | 2.5508 | 1.5685 | -1.3047 | -1.1775 | -1.1701 | -1.3002 | -1.1821 | -1.1678 | 6.9909 | |
| 2 | Trait | 2 | 1.5685 | 4.5539 | -1.4112 | -1.3534 | -0.9410 | -2.1592 | -2.1055 | -1.3149 | 6.9959 | |
| 3 | Trait*Animal | 1 | 1 | -1.3047 | -1.4112 | 1.8282 | 1.0652 | 1.0206 | 1.8010 | 1.0925 | 1.0070 | 0.05450 |
| 4 | Trait*Animal | 1 | 2 | -1.1775 | -1.3534 | 1.0652 | 1.7589 | 0.7085 | 1.0900 | 1.7341 | 0.7209 | -0.04955 |
| 5 | Trait*Animal | 1 | 3 | -1.1701 | -0.9410 | 1.0206 | 0.7085 | 1.7812 | 1.0095 | 0.7197 | 1.7756 | 0.02230 |
| 6 | Trait*Animal | 2 | 1 | -1.3002 | -2.1592 | 1.8010 | 1.0900 | 1.0095 | 2.7518 | 1.6392 | 1.4849 | 0.2651 |
| 7 | Trait*Animal | 2 | 2 | -1.1821 | -2.1055 | 1.0925 | 1.7341 | 0.7197 | 1.6392 | 2.6874 | 0.9930 | -0.2601 |
| 8 | Trait*Animal | 2 | 3 | -1.1678 | -1.3149 | 1.0070 | 0.7209 | 1.7756 | 1.4849 | 0.9930 | 2.7645 | 0.1276 |
The solutions for the fixed and random effects in Output 56.4.10 correspond to the last column in Output 56.4.9. Note that the standard errors for the fixed effects and the prediction standard errors for the random effects are the square root values of the diagonal entries in the solution of the mixed model equations (Output 56.4.9).
| Solution for Fixed Effects | ||||||
|---|---|---|---|---|---|---|
| Effect | Trait | Estimate | Standard Error | DF | t Value | Pr > |t| |
| Trait | 1 | 6.9909 | 1.5971 | 3 | 4.38 | 0.0221 |
| Trait | 2 | 6.9959 | 2.1340 | 3 | 3.28 | 0.0465 |
| Solution for Random Effects | |||||||
|---|---|---|---|---|---|---|---|
| Effect | Trait | Animal | Estimate | Std Err Pred | DF | t Value | Pr > |t| |
| Trait*Animal | 1 | 1 | 0.05450 | 1.3521 | 0 | 0.04 | . |
| Trait*Animal | 1 | 2 | -0.04955 | 1.3262 | 0 | -0.04 | . |
| Trait*Animal | 1 | 3 | 0.02230 | 1.3346 | 0 | 0.02 | . |
| Trait*Animal | 2 | 1 | 0.2651 | 1.6589 | 0 | 0.16 | . |
| Trait*Animal | 2 | 2 | -0.2601 | 1.6393 | 0 | -0.16 | . |
| Trait*Animal | 2 | 3 | 0.1276 | 1.6627 | 0 | 0.08 | . |
The estimates for the two traits are nearly identical, but the standard error of the second trait is larger because of the missing observation.
The Estimate column in the "Solution for Random Effects" table lists the best linear unbiased predictions (BLUPs) of the breeding values of both traits for all three animals. The p-values are missing because the default containment method for computing degrees of freedom results in zero degrees of freedom for the random effects parameter tests.
The two estimated traits are significantly different from zero at the 5% level (Output 56.4.11).
Output 56.4.12 displays the predicted values of the observations based on the trait and breeding value estimates—that is, the fixed and random effects.
| Obs | Trait | Animal | Y | Pred | StdErrPred | DF | Alpha | Lower | Upper | Resid |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 1 | 6 | 7.04542 | 1.33027 | 0 | 0.05 | . | . | -1.04542 |
| 2 | 1 | 2 | 8 | 6.94137 | 1.39806 | 0 | 0.05 | . | . | 1.05863 |
| 3 | 1 | 3 | 7 | 7.01321 | 1.41129 | 0 | 0.05 | . | . | -0.01321 |
| 4 | 2 | 1 | 9 | 7.26094 | 1.72839 | 0 | 0.05 | . | . | 1.73906 |
| 5 | 2 | 2 | 5 | 6.73576 | 1.74077 | 0 | 0.05 | . | . | -1.73576 |
| 6 | 2 | 3 | . | 7.12015 | 2.99088 | 0 | 0.05 | . | . | . |
The predicted values are not the predictions of future records in the sense that they do not contain a component corresponding to a new observational error. See Henderson (1984) for information about predicting future records. The Lower and Upper columns usually contain confidence limits for the predicted values; they are missing here because the random-effects parameter degrees of freedom equals 0.
Copyright © 2009 by SAS Institute Inc., Cary, NC, USA. All rights reserved.