| The CLUSTER Procedure |
Example 29.4 Evaluating the Effects of Ties
If, at some level of the cluster history, there is a tie for minimum distance between clusters, then one or more levels of the sample cluster tree are not uniquely determined. This example shows how the degree of indeterminacy can be assessed.
Mammals have four kinds of teeth: incisors, canines, premolars, and molars. The following data set gives the number of teeth of each kind on one side of the top and bottom jaws for 32 mammals.
Since all eight variables are measured in the same units, it is not strictly necessary to rescale the data. However, the canines have much less variance than the other kinds of teeth and, therefore, have little effect on the analysis if the variables are not standardized. An average linkage cluster analysis is run with and without standardization to enable comparison of the results.
title 'Hierarchical Cluster Analysis of Mammals'' Teeth Data';
title2 'Evaluating the Effects of Ties';
data teeth;
input mammal $ 1-16
@21 (v1-v8) (1.);
label v1='Top incisors'
v2='Bottom incisors'
v3='Top canines'
v4='Bottom canines'
v5='Top premolars'
v6='Bottom premolars'
v7='Top molars'
v8='Bottom molars';
datalines;
BROWN BAT 23113333
MOLE 32103333
SILVER HAIR BAT 23112333
PIGMY BAT 23112233
HOUSE BAT 23111233
RED BAT 13112233
PIKA 21002233
RABBIT 21003233
BEAVER 11002133
GROUNDHOG 11002133
GRAY SQUIRREL 11001133
HOUSE MOUSE 11000033
PORCUPINE 11001133
WOLF 33114423
BEAR 33114423
RACCOON 33114432
MARTEN 33114412
WEASEL 33113312
WOLVERINE 33114412
BADGER 33113312
RIVER OTTER 33114312
SEA OTTER 32113312
JAGUAR 33113211
COUGAR 33113211
FUR SEAL 32114411
SEA LION 32114411
GREY SEAL 32113322
ELEPHANT SEAL 21114411
REINDEER 04103333
ELK 04103333
DEER 04003333
MOOSE 04003333
;
The following statements produce Output 29.4.1:
title3 'Raw Data';
proc cluster data=teeth method=average nonorm noeigen;
var v1-v8;
id mammal;
run;
| Root-Mean-Square Total-Sample Standard Deviation | 0.898027 |
|---|
| Cluster History | |||||
|---|---|---|---|---|---|
| NCL | Clusters Joined | FREQ | RMS Dist |
T i e |
|
| 31 | BEAVER | GROUNDHOG | 2 | 0 | T |
| 30 | GRAY SQUIRREL | PORCUPINE | 2 | 0 | T |
| 29 | WOLF | BEAR | 2 | 0 | T |
| 28 | MARTEN | WOLVERINE | 2 | 0 | T |
| 27 | WEASEL | BADGER | 2 | 0 | T |
| 26 | JAGUAR | COUGAR | 2 | 0 | T |
| 25 | FUR SEAL | SEA LION | 2 | 0 | T |
| 24 | REINDEER | ELK | 2 | 0 | T |
| 23 | DEER | MOOSE | 2 | 0 | |
| 22 | BROWN BAT | SILVER HAIR BAT | 2 | 1 | T |
| 21 | PIGMY BAT | HOUSE BAT | 2 | 1 | T |
| 20 | PIKA | RABBIT | 2 | 1 | T |
| 19 | CL31 | CL30 | 4 | 1 | T |
| 18 | CL28 | RIVER OTTER | 3 | 1 | T |
| 17 | CL27 | SEA OTTER | 3 | 1 | T |
| 16 | CL24 | CL23 | 4 | 1 | |
| 15 | CL21 | RED BAT | 3 | 1.2247 | |
| 14 | CL17 | GREY SEAL | 4 | 1.291 | |
| 13 | CL29 | RACCOON | 3 | 1.4142 | T |
| 12 | CL25 | ELEPHANT SEAL | 3 | 1.4142 | |
| 11 | CL18 | CL14 | 7 | 1.5546 | |
| 10 | CL22 | CL15 | 5 | 1.5811 | |
| 9 | CL20 | CL19 | 6 | 1.8708 | T |
| 8 | CL11 | CL26 | 9 | 1.9272 | |
| 7 | CL8 | CL12 | 12 | 2.2278 | |
| 6 | MOLE | CL13 | 4 | 2.2361 | |
| 5 | CL9 | HOUSE MOUSE | 7 | 2.4833 | |
| 4 | CL6 | CL7 | 16 | 2.5658 | |
| 3 | CL10 | CL16 | 9 | 2.8107 | |
| 2 | CL3 | CL5 | 16 | 3.7054 | |
| 1 | CL2 | CL4 | 32 | 4.2939 | |
The following statements produce Output 29.4.2:
title3 'Standardized Data';
proc cluster data=teeth std method=average nonorm noeigen;
var v1-v8;
id mammal;
run;
| Root-Mean-Square Total-Sample Standard Deviation | 1 |
|---|
| Cluster History | |||||
|---|---|---|---|---|---|
| NCL | Clusters Joined | FREQ | RMS Dist |
T i e |
|
| 31 | BEAVER | GROUNDHOG | 2 | 0 | T |
| 30 | GRAY SQUIRREL | PORCUPINE | 2 | 0 | T |
| 29 | WOLF | BEAR | 2 | 0 | T |
| 28 | MARTEN | WOLVERINE | 2 | 0 | T |
| 27 | WEASEL | BADGER | 2 | 0 | T |
| 26 | JAGUAR | COUGAR | 2 | 0 | T |
| 25 | FUR SEAL | SEA LION | 2 | 0 | T |
| 24 | REINDEER | ELK | 2 | 0 | T |
| 23 | DEER | MOOSE | 2 | 0 | |
| 22 | PIGMY BAT | RED BAT | 2 | 0.9157 | |
| 21 | CL28 | RIVER OTTER | 3 | 0.9169 | |
| 20 | CL31 | CL30 | 4 | 0.9428 | T |
| 19 | BROWN BAT | SILVER HAIR BAT | 2 | 0.9428 | T |
| 18 | PIKA | RABBIT | 2 | 0.9428 | |
| 17 | CL27 | SEA OTTER | 3 | 0.9847 | |
| 16 | CL22 | HOUSE BAT | 3 | 1.1437 | |
| 15 | CL21 | CL17 | 6 | 1.3314 | |
| 14 | CL25 | ELEPHANT SEAL | 3 | 1.3447 | |
| 13 | CL19 | CL16 | 5 | 1.4688 | |
| 12 | CL15 | GREY SEAL | 7 | 1.6314 | |
| 11 | CL29 | RACCOON | 3 | 1.692 | |
| 10 | CL18 | CL20 | 6 | 1.7357 | |
| 9 | CL12 | CL26 | 9 | 2.0285 | |
| 8 | CL24 | CL23 | 4 | 2.1891 | |
| 7 | CL9 | CL14 | 12 | 2.2674 | |
| 6 | CL10 | HOUSE MOUSE | 7 | 2.317 | |
| 5 | CL11 | CL7 | 15 | 2.6484 | |
| 4 | CL13 | MOLE | 6 | 2.8624 | |
| 3 | CL4 | CL8 | 10 | 3.5194 | |
| 2 | CL3 | CL6 | 17 | 4.1265 | |
| 1 | CL2 | CL5 | 32 | 4.7753 | |
There are ties at 16 levels for the raw data but at only 10 levels for the standardized data. There are more ties for the raw data because the increments between successive values are the same for all of the raw variables but different for the standardized variables.
One way to assess the importance of the ties in the analysis is to repeat the analysis on several random permutations of the observations and then to see to what extent the results are consistent at the interesting levels of the cluster history. Three macros are presented to facilitate this process, as follows.
/* --------------------------------------------------------- */
/* */
/* The macro CLUSPERM randomly permutes observations and */
/* does a cluster analysis for each permutation. */
/* The arguments are as follows: */
/* */
/* data data set name */
/* var list of variables to cluster */
/* id id variable for proc cluster */
/* method clustering method (and possibly other options) */
/* nperm number of random permutations. */
/* */
/* --------------------------------------------------------- */
%macro CLUSPERM(data,var,id,method,nperm);
/* ------CREATE TEMPORARY DATA SET WITH RANDOM NUMBERS------ */
data _temp_;
set &data;
array _random_ _ran_1-_ran_&nperm;
do over _random_;
_random_=ranuni(835297461);
end;
run;
/* ------PERMUTE AND CLUSTER THE DATA----------------------- */
%do n=1 %to &nperm;
proc sort data=_temp_(keep=_ran_&n &var &id) out=_perm_;
by _ran_&n;
run;
proc cluster method=&method noprint outtree=_tree_&n;
var &var;
id &id;
run;
%end;
%mend;
/* --------------------------------------------------------- */
/* */
/* The macro PLOTPERM plots various cluster statistics */
/* against the number of clusters for each permutation. */
/* The arguments are as follows: */
/* */
/* nclus maximum number of clusters to be plotted */
/* nperm number of random permutations. */
/* */
/* --------------------------------------------------------- */
%macro PLOTPERM(nclus,nperm);
/* ---CONCATENATE TREE DATA SETS FOR 20 OR FEWER CLUSTERS--- */
data _plot_;
set %do n=1 %to &nperm; _tree_&n(in=_in_&n) %end; ;
if _ncl_<=&nclus;
%do n=1 %to &nperm;
if _in_&n then _perm_=&n;
%end;
label _perm_='permutation number';
keep _ncl_ _psf_ _pst2_ _ccc_ _perm_;
run;
/* ---PLOT THE REQUESTED STATISTICS BY NUMBER OF CLUSTERS--- */
proc sgscatter ;
compare y=(_ccc_ _psf_ _pst2_) x=_ncl_ /group=_perm_ ;
run;
%mend;
/* --------------------------------------------------------- */
/* */
/* The macro TABPERM generates cluster-membership variables */
/* for a specified number of clusters for each permutation. */
/* PROC TABULATE gives the frequencies and means. */
/* The arguments are as follows: */
/* */
/* var list of variables to cluster */
/* (no "-" or ":" allowed) */
/* id id variable for proc cluster */
/* meanfmt format for printing means in PROC TABULATE */
/* nclus number of clusters desired */
/* nperm number of random permutations. */
/* */
/* --------------------------------------------------------- */
%macro TABPERM(var,id,meanfmt,nclus,nperm);
/* ------CREATE DATA SETS GIVING CLUSTER MEMBERSHIP--------- */
%do n=1 %to &nperm;
proc tree data=_tree_&n noprint n=&nclus
out=_out_&n(drop=clusname
rename=(cluster=_clus_&n));
copy &var;
id &id;
run;
proc sort;
by &id &var;
run;
%end;
/* ------MERGE THE CLUSTER VARIABLES------------------------ */
data _merge_;
merge
%do n=1 %to &nperm;
_out_&n
%end; ;
by &id &var;
length all_clus $ %eval(3*&nperm);
%do n=1 %to &nperm;
substr( all_clus, %eval(1+(&n-1)*3), 3) =
put( _clus_&n, 3.);
%end;
run;
/* ------ TABULATE CLUSTER COMBINATIONS------------ */
proc sort;
by _clus_:;
run;
proc tabulate order=data formchar=' ';
class all_clus;
var &var;
table all_clus, n='FREQ'*f=5. mean*f=&meanfmt*(&var) /
rts=%eval(&nperm*3+1);
run;
%mend;
To use these macros, it is first convenient to define a macro, VLIST, listing the teeth variables, since the forms V1-V8 or V: cannot be used with the TABULATE procedure in the TABPERM macro:
/* -TABULATE does not accept hyphens or colons in VAR lists- */ %let vlist=v1 v2 v3 v4 v5 v6 v7 v8;
The CLUSPERM macro is then called to analyze 10 random permutations. The PLOTPERM macro plots the pseudo 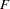 and
and 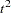 statistics and the cubic clustering criterion. Since the data are discrete, the pseudo
statistics and the cubic clustering criterion. Since the data are discrete, the pseudo 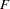 statistic and the cubic clustering criterion can be expected to increase as the number of clusters increases, so local maxima or large jumps in these statistics are more relevant than the global maximum in determining the number of clusters. For the raw data, only the pseudo
statistic and the cubic clustering criterion can be expected to increase as the number of clusters increases, so local maxima or large jumps in these statistics are more relevant than the global maximum in determining the number of clusters. For the raw data, only the pseudo 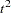 statistic indicates the possible presence of clusters, with the 4-cluster level being suggested. Hence, the macros are used as follows to analyze the results at the 4-cluster level:
statistic indicates the possible presence of clusters, with the 4-cluster level being suggested. Hence, the macros are used as follows to analyze the results at the 4-cluster level:
title3 'Raw Data'; /* ------CLUSTER RAW DATA WITH AVERAGE LINKAGE-------------- */ %clusperm( teeth, &vlist, mammal, average, 10);
The following statements produce Output 29.4.3.
/* -----PLOT STATISTICS FOR THE LAST 20 LEVELS-------------- */ %plotperm(20, 10);
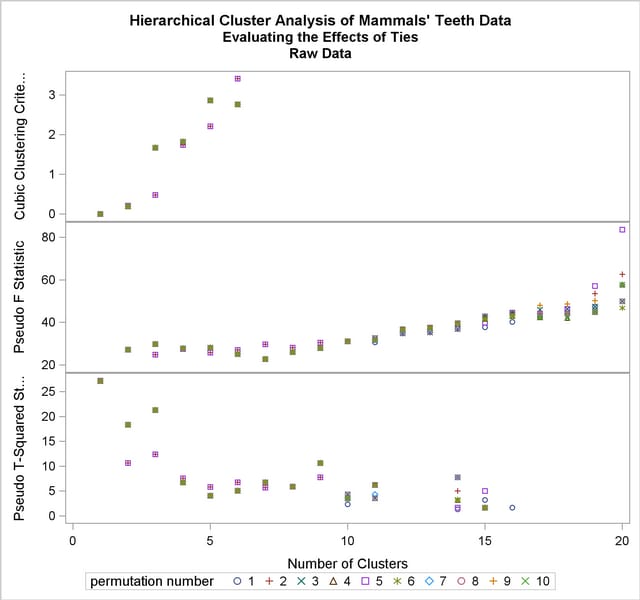
The following statements produce Output 29.4.4.
/* ------ANALYZE THE 4-CLUSTER LEVEL------------------------ */ %tabperm( &vlist, mammal, 9.1, 4, 10);
| Hierarchical Cluster Analysis of Mammals' Teeth Data |
| Evaluating the Effects of Ties |
| Raw Data |
| FREQ | Mean | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Top incisors | Bottom incisors | Top canines | Bottom canines | Top premolars | Bottom premolars | Top molars | Bottom molars | ||
| all_clus | 4 | 0.0 | 4.0 | 0.5 | 0.0 | 3.0 | 3.0 | 3.0 | 3.0 |
| 1 3 1 1 1 3 3 3 2 3 | |||||||||
| 2 2 2 2 2 2 1 2 1 1 | 15 | 2.9 | 2.6 | 1.0 | 1.0 | 3.6 | 3.4 | 1.3 | 1.8 |
| 2 4 2 2 4 2 1 2 1 1 | 1 | 3.0 | 2.0 | 1.0 | 0.0 | 3.0 | 3.0 | 3.0 | 3.0 |
| 3 1 3 3 3 1 2 1 3 2 | 5 | 1.0 | 1.0 | 0.0 | 0.0 | 1.2 | 0.8 | 3.0 | 3.0 |
| 3 4 3 3 4 1 2 1 3 2 | 2 | 2.0 | 1.0 | 0.0 | 0.0 | 2.5 | 2.0 | 3.0 | 3.0 |
| 4 4 4 4 4 4 4 4 4 4 | 5 | 1.8 | 3.0 | 1.0 | 1.0 | 2.0 | 2.4 | 3.0 | 3.0 |
From the TABULATE output, you can see that two types of clustering are obtained. In one case, the mole is grouped with the carnivores, while the pika and rabbit are grouped with the rodents. In the other case, both the mole and the lagomorphs are grouped with the bats.
Next, the analysis is repeated with the standardized data as shown in the following statements. The pseudo 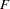 and
and 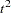 statistics indicate 3 or 4 clusters, while the cubic clustering criterion shows a sharp rise up to 4 clusters and then levels off up to 6 clusters. So the TABPERM macro is used again at the 4-cluster level. In this case, there is no indeterminacy, because the same four clusters are obtained with every permutation, although in different orders. It must be emphasized, however, that lack of indeterminacy in no way indicates validity.
statistics indicate 3 or 4 clusters, while the cubic clustering criterion shows a sharp rise up to 4 clusters and then levels off up to 6 clusters. So the TABPERM macro is used again at the 4-cluster level. In this case, there is no indeterminacy, because the same four clusters are obtained with every permutation, although in different orders. It must be emphasized, however, that lack of indeterminacy in no way indicates validity.
title3 'Standardized Data'; /*------CLUSTER STANDARDIZED DATA WITH AVERAGE LINKAGE------*/ %clusperm( teeth, &vlist, mammal, average std, 10);
The following statements produce Output 29.4.5.
/* -----PLOT STATISTICS FOR THE LAST 20 LEVELS-------------- */ %plotperm(20, 10);
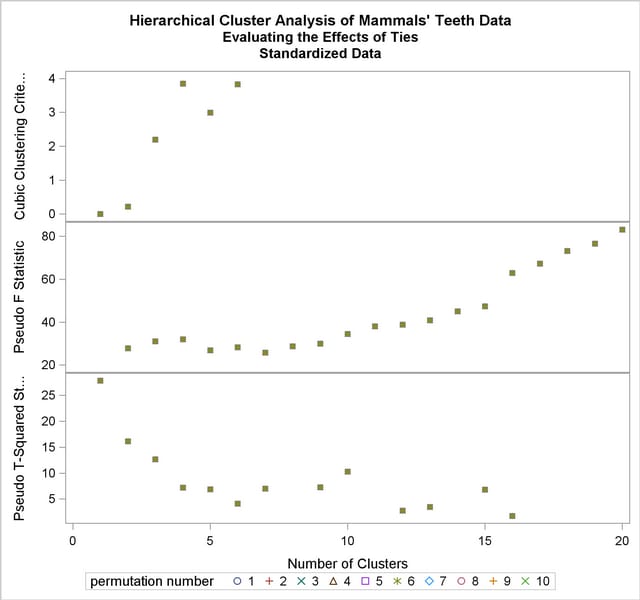
The following statements produce Output 29.4.6.
/* ------ANALYZE THE 4-CLUSTER LEVEL------------------------ */ %tabperm( &vlist, mammal, 9.1, 4, 10);
| Hierarchical Cluster Analysis of Mammals' Teeth Data |
| Evaluating the Effects of Ties |
| Standardized Data |
| FREQ | Mean | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Top incisors | Bottom incisors | Top canines | Bottom canines | Top premolars | Bottom premolars | Top molars | Bottom molars | ||
| all_clus | 4 | 0.0 | 4.0 | 0.5 | 0.0 | 3.0 | 3.0 | 3.0 | 3.0 |
| 1 3 1 1 1 3 3 3 2 3 | |||||||||
| 2 2 2 2 2 2 1 2 1 1 | 15 | 2.9 | 2.6 | 1.0 | 1.0 | 3.6 | 3.4 | 1.3 | 1.8 |
| 3 1 3 3 3 1 2 1 3 2 | 7 | 1.3 | 1.0 | 0.0 | 0.0 | 1.6 | 1.1 | 3.0 | 3.0 |
| 4 4 4 4 4 4 4 4 4 4 | 6 | 2.0 | 2.8 | 1.0 | 0.8 | 2.2 | 2.5 | 3.0 | 3.0 |
Copyright © 2009 by SAS Institute Inc., Cary, NC, USA. All rights reserved.