-
PREDICTED<(COMPONENT | OVERALL)>
PRED<(COMPONENT | OVERALL)>
MEAN<(COMPONENT | OVERALL)>
-
requests predicted values (predicted means) for the response variable. The predictions in the output data set are mapped onto
the data scale in all cases except for a binomial or binary response with events/trials syntax and when PREDTYPE=COUNT
has not been specified. In that case the predictions are predicted success probabilities.
The default is to compute the predicted value for the mixture (OVERALL). You can request predictions for the means of the
component distributions by adding the COMPONENT suboption in parentheses. The predicted values for some distributions are
not identical to the parameter modeled as 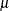 . For example, in the lognormal distribution the predicted mean is
. For example, in the lognormal distribution the predicted mean is 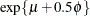 where
where 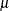 and
and  are the parameters of an underlying normal process; see the section Log-Likelihood Functions for Response Distributions for details.
are the parameters of an underlying normal process; see the section Log-Likelihood Functions for Response Distributions for details.
-
RESIDUAL<(COMPONENT | OVERALL)>
RESID<(COMPONENT | OVERALL)>
-
requests residuals for the response or residuals in the component distributions. Only "raw" residuals on the data scale are
computed (observed minus predicted).
-
VARIANCE<(COMPONENT | OVERALL)>
VAR<(COMPONENT | OVERALL)>
-
requests variances for the mixture or the component distributions.
-
LOGLIKE<(COMPONENT | OVERALL)>
LOGL<(COMPONENT | OVERALL)>
-
requests values of the log-likelihood function for the mixture or the components. For observations used in the analysis, the
overall computed value is the observations’ contribution to the log likelihood; if a FREQ
statement is present, the frequency is accounted for in the computed value. In other words, if all observations in the input
data set have been used in the analysis, adding the value of the log-likelihood contributions in the OUTPUT data set produces
the negative of the final objective function value in the "Iteration History" table. By default, the log-likelihood contribution
to the mixture is computed. You can request the individual mixture component contributions with the COMPONENT suboption.
-
MIXPROBS<(COMPONENT | MAX)>
MIXPROB<(COMPONENT | MAX)>
PRIOR<(COMPONENT | MAX)>
MIXWEIGHTS<(COMPONENT | MAX)>
-
requests that the prior weights 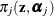 be added to the OUTPUT data set. By default, the probabilities are output for all components. You can limit the output to
a single statistic, the largest mixing probability, with the MAX suboption.
be added to the OUTPUT data set. By default, the probabilities are output for all components. You can limit the output to
a single statistic, the largest mixing probability, with the MAX suboption.
Note: The keyword "prior" is used here because of long-standing practice to refer to the mixing probabilities as prior weights.
This must not be confused with the prior distribution and its parameters in a Bayesian analysis.
-
POSTERIOR<(COMPONENT | MAX)>
POST<(COMPONENT | MAX)>
PROB<(COMPONENT | MAX)>
-
requests that the posterior weights
be added to the OUTPUT data set. By default, the probabilities are output for all components. You can limit the output to
a single statistic, the largest posterior probability, with the MAX suboption.
Note: The keyword "posterior" is used here because of long-standing practice to refer to these probabilities as posterior probabilities.
This must not be confused with the posterior distribution in a Bayesian analysis.
-
LINP
XBETA
-
requests that the linear predictors for the models be added to the OUTPUT data set.
-
CLASS | CATEGORY | GROUP
-
adds the estimated component membership to the OUTPUT data set. An observation is associated with the component that has the
highest posterior probability.
-
MAXPOST | MAXPROB
-
adds the highest posterior probability to the OUTPUT data set.

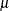
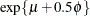

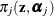
![\[ \frac{\pi _ j(\mb{z},\balpha _ j) p_ j(y;\mb{x}_ j'\bbeta _ j,\phi _ j)}{\sum _{j=1}^{k} \pi _ j(\mb{z},\balpha _ j) p_ j(y;\mb{x}_ j'\bbeta _ j,\phi _ j)} \]](images/stathpug_hpfmm0132.png)