The MODEL statement defines the statistical model in terms of a response variable (the target) or an events/trials specification. You can also specify model effects that are constructed from variables in the input data set, and you can
specify options. An intercept is included in the model by default. You can remove the intercept by specifying the NOINT option.
You can specify a single response variable that contains your interval, binary, ordinal, or nominal response values. When you have binomial data, you can specify
the events/trials form of the response, where one variable contains the number of positive responses (or events) and another variable contains
the number of trials. The values of both events and (trials – events) must be nonnegative, and the value of trials must be positive. If you specify a single response variable that is in a CLASS
statement, then the response is assumed to be either binary or multinomial, depending on the number of levels.
For information about constructing the model effects, see the section Specification and Parameterization of Model Effects of Chapter 4: Shared Statistical Concepts.
There are two sets of options in the MODEL statement. The response-options
determine how the HPGENSELECT procedure models probabilities for binary and multinomial data. The model-options
control other aspects of model formation and inference. Table 7.3 summarizes these options.
Table 7.3: MODEL Statement Options
|
Option
|
Description
|
|
Response Variable Options for Binary and Multinomial Models
|
|
DESCENDING
|
Reverses the response categories
|
|
EVENT=
|
Specifies the event category
|
|
ORDER=
|
Specifies the sort order
|
|
REF=
|
Specifies the reference category
|
|
Model Options
|
|
ALPHA=
|
Specifies the confidence level for confidence limits
|
|
CL
|
Requests confidence limits
|
|
DISPERSION | PHI=
|
Specifies a fixed dispersion parameter
|
|
DISTRIBUTION | DIST=
|
Specifies the response distribution
|
|
INCLUDE=
|
Includes effects in all models for model selection
|
|
INITIALPHI=
|
Specifies a starting value of the dispersion parameter
|
|
LINK=
|
Specifies the link function
|
|
NOCENTER
|
Requests that continuous main effects not be centered and scaled
|
|
NOINT
|
Suppresses the intercept
|
|
OFFSET=
|
Specifies the offset variable
|
|
SAMPLEFRAC=
|
Specifies the fraction of the data to be used to compute starting
|
| |
values for the Tweedie distribution
|
|
START=
|
Includes effects in the initial model for model selection
|
Response Variable Options
Response variable options determine how the HPGENSELECT procedure models probabilities for binary and multinomial data.
You can specify the following response-options by enclosing them in parentheses after the response or trials variable.
-
DESCENDING
DESC
-
reverses the order of the response categories. If both the DESCENDING and ORDER=
options are specified, PROC HPGENSELECT orders the response categories according to the ORDER=
option and then reverses that order.
-
EVENT=’category’ | FIRST | LAST
-
specifies the event category for the binary response model. PROC HPGENSELECT models the probability of the event category.
The EVENT= option has no effect when there are more than two response categories.
You can specify the event category (formatted, if a format is applied) in quotes, or you can specify one of the following:
-
FIRST
-
designates the first ordered category as the event. This is the default.
-
LAST
-
designates the last ordered category as the event.
For example, the following statements specify that observations that have a formatted value of '1' represent events in the
data. The probability modeled by the HPGENSELECT procedure is thus the probability that the variable def takes on the (formatted) value '1'.
proc hpgenselect data=MyData;
class A B C;
model def(event ='1') = A B C x1 x2 x3;
run;
-
ORDER=DATA | FORMATTED | INTERNAL
ORDER=FREQ | FREQDATA | FREQFORMATTED | FREQINTERNAL
-
specifies the sort order for the levels of the response variable. When ORDER=FORMATTED (the default) for numeric variables for which you have supplied no explicit format (that is,
for which there is no corresponding FORMAT statement in the current PROC HPGENSELECT run or in the DATA step that created
the data set), the levels are ordered by their internal (numeric) value. Table 7.4 shows the interpretation of the ORDER= option.
Table 7.4: Sort Order
|
ORDER=
|
Levels Sorted By
|
|
DATA
|
Order of appearance in the input data set
|
|
FORMATTED
|
External formatted value, except for numeric variables that have no explicit format, which are sorted by their unformatted
(internal) value
|
|
FREQ
|
Descending frequency count (levels that have the most observations come first in the order)
|
|
FREQDATA
|
Order of descending frequency count; within counts by order of appearance in the input data set when counts are tied
|
|
FREQFORMATTED
|
Order of descending frequency count; within counts by formatted value when counts are tied
|
|
FREQINTERNAL
|
Order of descending frequency count; within counts by unformatted value when counts are tied
|
|
INTERNAL
|
Unformatted value
|
By default, ORDER=FORMATTED. For the FORMATTED and INTERNAL orders, the sort order is machine-dependent.
For more information about sort order, see the chapter about the SORT procedure in Base SAS Procedures Guide and the discussion of BY-group processing in SAS Language Reference: Concepts.
-
REF=’category’ | FIRST | LAST
-
specifies the reference category for the generalized logit model and the binary response model. For the generalized logit
model, each logit contrasts a nonreference category with the reference category. For the binary response model, specifying
one response category as the reference is the same as specifying the other response category as the event category. You can
specify the reference category (formatted if a format is applied) in quotes, or you can specify one of the following:
-
FIRST
-
designates the first ordered category as the reference
-
LAST
-
designates the last ordered category as the reference. This is the default.
-
ALPHA=number
-
requests that confidence intervals for each of the parameters that are requested by the CL
option be constructed with confidence level 1–number. The value of number must be between 0 and 1; the default is 0.05.
-
CL
-
requests that confidence limits be constructed for each of the parameter estimates. The confidence level is 0.95 by default;
this can be changed by specifying the ALPHA=
option.
-
DISPERSION=number
-
specifies a fixed dispersion parameter for those distributions that have a dispersion parameter. The dispersion parameter
used in all computations is fixed at number, and not estimated.
-
DISTRIBUTION=keyword
-
specifies the response distribution for the model. The keywords and the associated distributions are shown in Table 7.5.
Table 7.5: Built-In Distribution Functions
| |
Distribution
|
|
DISTRIBUTION=
|
Function
|
|
BINARY
|
Binary
|
|
BINOMIAL
|
Binary or binomial
|
|
GAMMA
|
Gamma
|
|
INVERSEGAUSSIAN | IG
|
Inverse Gaussian
|
|
MULTINOMIAL | MULT
|
Multinomial
|
|
NEGATIVEBINOMIAL | NB
|
Negative binomial
|
|
NORMAL | GAUSSIAN
|
Normal
|
|
POISSON
|
Poisson
|
|
TWEEDIE<(Tweedie-options)>
|
Tweedie
|
|
ZINB
|
Zero-inflated negative binomial
|
|
ZIP
|
Zero-inflated Poisson
|
When DISTRIBUTION=TWEEDIE, you can specify the following Tweedie-options:
-
INITIALP=
-
specifies a starting value for iterative estimation of the Tweedie power parameter.
-
OPTMETHOD=Tweedie-optimization-option
-
requests an optimization method for iterative estimation of the Tweedie model parameters. You can specify the following Tweedie-optimization-options:
-
EQL
-
requests that extended quasi-likelihood be used for a sample of the data, followed by extended quasi-likelihood for the full
data. This is equivalent to the TWEEDIEEQL Tweedie-option.
-
EQLLHOOD
-
requests that extended quasi-likelihood be used for a sample of the data, followed by Tweedie log likelihood for the full
data. This is the default method.
-
FINALLHOOD
-
requests a four-stage approach to estimating the Tweedie model parameters. The four stages are as follows:
-
extended quasi-likelihood for a sample of the data
-
Tweedie log likelihood for a sample of the data
-
extended quasi-likelihood for the full data
-
Tweedie log likelihood for the full data
-
LHOOD
-
requests that Tweedie log likelihood be used for a sample of the data, followed by Tweedie log likelihood for the full data.
-
P=
-
requests a fixed Tweedie power parameter.
-
TWEEDIEEQL | EQL
-
requests that extended quasi-likelihood be used instead of Tweedie log likelihood in parameter estimation.
If you do not specify a link function with the LINK=
option, a default link function is used. The default link function for each distribution is shown in Table 7.6. For the binary and multinomial distributions, only the link functions shown in Table 7.6 are available. For the other distributions, you can use any link function shown in Table 7.7 by specifying the LINK=
option. Other commonly used link functions for each distribution are shown in Table 7.6.
Table 7.6: Default and Commonly Used Link Functions
| |
Default
|
Other Commonly Used
|
|
DISTRIBUTION=
|
Link Function
|
Link Functions
|
|
BINARY
|
Logit
|
Probit, complementary log-log, log-log
|
|
BINOMIAL
|
Logit
|
Probit, complementary log-log, log-log
|
|
GAMMA
|
Reciprocal
|
Log
|
|
INVERSEGAUSSIAN | IG
|
Reciprocal square
|
Log
|
|
MULTINOMIAL | MULT
|
Logit (ordinal)
|
Probit, complementary log-log, log-log
|
|
MULTINOMIAL | MULT
|
Generalized logit (nominal)
|
|
|
NEGATIVEBINOMIAL | NB
|
Log
|
|
|
NORMAL | GAUSSIAN
|
Identity
|
Log
|
|
POISSON
|
Log
|
|
|
TWEEDIE
|
Log
|
|
|
ZINB
|
Log
|
|
|
ZIP
|
Log
|
|
-
INCLUDE=n
INCLUDE=single-effect
INCLUDE=(effects)
-
forces effects to be included in all models. If you specify INCLUDE=n, then the first n effects that are listed in the MODEL
statement are included in all models. If you specify INCLUDE=single-effect or if you specify a list of effects within parentheses, then the specified effects are forced into all models. The effects
that you specify in this option must be explanatory effects that are specified in the MODEL
statement before the slash (/).
-
INITIAL-PHI=number
-
specifies a starting value for iterative maximum likelihood estimation of the dispersion parameter for distributions that
have a dispersion parameter.
-
LINK=keyword
-
specifies the link function for the model. The keywords and the associated link functions are shown in Table 7.7. Default and commonly used link functions for the available distributions are shown in Table 7.6.
Table 7.7: Built-In Link Functions
| |
Link
|
|
|
LINK=
|
Function
|
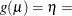
|
|
CLOGLOG | CLL
|
Complementary log-log
|
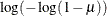
|
|
GLOGIT | GENLOGIT
|
Generalized logit
|
|
|
IDENTITY | ID
|
Identity
|
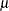
|
|
INV | RECIP
|
Reciprocal
|

|
|
INV2
|
Reciprocal square
|

|
|
LOG
|
Logarithm
|

|
|
LOGIT
|
Logit
|
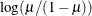
|
|
LOGLOG
|
Log-log
|
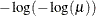
|
|
PROBIT
|
Probit
|
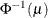
|
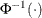 denotes the quantile function of the standard normal distribution.
denotes the quantile function of the standard normal distribution.
If a multinomial response variable has more than two categories, the HPGENSELECT procedure fits a model by using a cumulative
link function that is based on the specified link. However, if you specify LINK=GLOGIT, the procedure assumes a generalized
logit model for nominal (unordered) data, regardless of the number of response categories.
-
NOCENTER
-
requests that continuous main effects not be centered and scaled internally. (Continuous main effects are centered and scaled
by default to aid in computing maximum likelihood estimates.) Parameter estimates and related statistics are always reported
on the original scale.
-
NOINT
-
requests that no intercept be included in the model. (An intercept is included by default.) The NOINT option is not available
in multinomial models.
-
OFFSET=variable
-
specifies a variable to be used as an offset to the linear predictor. An offset plays the role of an effect whose coefficient is known to be 1.
The offset variable cannot appear in the CLASS
statement or elsewhere in the MODEL
statement. Observations that have missing values for the offset variable are excluded from the analysis.
-
SAMPLEFRAC=number
-
specifies a fraction of the data to be used to determine starting values for iterative estimation of the parameters of a Tweedie
model. The sampled data are used in an extended quasi-likelihood estimation of the model parameters. The estimated parameters
are then used as starting values in a full maximum likelihood estimation of the model parameters that uses all of the data.
-
START=n
START=single-effect
START=(effects)
-
begins the selection process from the designated initial model for the FORWARD and STEPWISE selection methods. If you specify
START=n, then the starting model includes the first n effects that are listed in the MODEL
statement. If you specify START=single-effect or if you specify a list of effects within parentheses, then the starting model includes those specified effects. The effects
that you specify in the START= option must be explanatory effects that are specified in the MODEL
statement before the slash (/). The START= option is not available when you specify METHOD=BACKWARD in the SELECTION
statement.
