-
ALPHA
-
calculates and prints Cronbach’s coefficient alpha. PROC CORR computes separate coefficients using raw and standardized values
(scaling the variables to a unit variance of 1). For each VAR statement variable, PROC CORR computes the correlation between
the variable and the total of the remaining variables. It also computes Cronbach’s coefficient alpha by using only the remaining
variables.
If a WITH statement is specified, the ALPHA option is invalid. When you specify the ALPHA option, the Pearson correlations
will also be displayed. If you specify the OUTP= option, the output data set also contains observations with Cronbach’s coefficient
alpha. If you use the PARTIAL statement, PROC CORR calculates Cronbach’s coefficient alpha for partialled variables. See the
section Partial Correlation for details.
-
BEST=n
-
prints the n highest correlation coefficients for each variable, 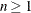 . Correlations are ordered from highest to lowest in absolute value. Otherwise, PROC CORR prints correlations in a rectangular
table, using the variable names as row and column labels.
. Correlations are ordered from highest to lowest in absolute value. Otherwise, PROC CORR prints correlations in a rectangular
table, using the variable names as row and column labels.
If you specify the HOEFFDING option, PROC CORR displays the 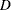 statistics in order from highest to lowest.
statistics in order from highest to lowest.
-
COV
-
displays the variance and covariance matrix. When you specify the COV option, the Pearson correlations will also be displayed.
If you specify the OUTP= option, the output data set also contains the covariance matrix with the corresponding _TYPE_ variable
value 'COV.' If you use the PARTIAL statement, PROC CORR computes a partial covariance matrix.
-
CSSCP
-
displays a table of the corrected sums of squares and crossproducts. When you specify the CSSCP option, the Pearson correlations
will also be displayed. If you specify the OUTP= option, the output data set also contains a CSSCP matrix with the corresponding
_TYPE_ variable value 'CSSCP.' If you use a PARTIAL statement, PROC CORR prints both an unpartial and a partial CSSCP matrix,
and the output data set contains a partial CSSCP matrix.
-
DATA=SAS-data-set
-
names the SAS data set to be analyzed by PROC CORR. By default, the procedure uses the most recently created SAS data set.
-
EXCLNPWGT
EXCLNPWGTS
-
excludes observations with nonpositive weight values from the analysis. By default, PROC CORR treats observations with negative
weights like those with zero weights and counts them in the total number of observations.
-
FISHER <(fisher-options)>
-
requests confidence limits and p-values under a specified null hypothesis, 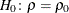 , for correlation coefficients by using Fisher’s z transformation. These correlations include the Pearson correlations and Spearman correlations.
, for correlation coefficients by using Fisher’s z transformation. These correlations include the Pearson correlations and Spearman correlations.
The following fisher-options are available:
-
HOEFFDING
-
requests a table of Hoeffding’s D statistics. This D statistic is 30 times larger than the usual definition and scales the range between –0.5 and 1 so that large positive values
indicate dependence. The HOEFFDING option is invalid if a WEIGHT or PARTIAL statement is used.
-
KENDALL
-
requests a table of Kendall’s tau-b coefficients based on the number of concordant and discordant pairs of observations. Kendall’s
tau-b ranges from –1 to 1.
The KENDALL option is invalid if a WEIGHT statement is used. If you use a PARTIAL statement, probability values for Kendall’s
partial tau-b are not available.
-
NOCORR
-
suppresses displaying of Pearson correlations. If you specify the OUTP= option, the data set type remains CORR. To change
the data set type to COV, CSSCP, or SSCP, use the TYPE= data set option.
-
NOMISS
-
excludes observations with missing values from the analysis. Otherwise, PROC CORR computes correlation statistics by using
all of the nonmissing pairs of variables. Using the NOMISS option is computationally more efficient.
-
NOPRINT
-
suppresses all displayed output, which also includes output produced with ODS Graphics. Use the NOPRINT option if you want
to create an output data set only.
-
NOPROB
-
suppresses displaying the probabilities associated with each correlation coefficient.
-
NOSIMPLE
-
suppresses printing simple descriptive statistics for each variable. However, if you request an output data set, the output
data set still contains simple descriptive statistics for the variables.
-
OUTH=output-data-set
-
creates an output data set that contains Hoeffding’s 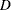 statistics. The contents of the output data set are similar to those of the OUTP= data set. When you specify the OUTH= option,
the Hoeffding’s
statistics. The contents of the output data set are similar to those of the OUTP= data set. When you specify the OUTH= option,
the Hoeffding’s 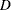 statistics will be displayed.
statistics will be displayed.
-
OUTK=output-data-set
-
creates an output data set that contains Kendall correlation statistics. The contents of the output data set are similar to
those of the OUTP= data set. When you specify the OUTK= option, the Kendall correlation statistics will be displayed.
-
OUTP=output-data-set
OUT=output-data-set
-
creates an output data set that contains Pearson correlation statistics. This data set also includes means, standard deviations,
and the number of observations. The value of the _TYPE_ variable is 'CORR.' When you specify the OUTP= option, the Pearson
correlations will also be displayed. If you specify the ALPHA option, the output data set also contains six observations with
Cronbach’s coefficient alpha.
-
OUTPLC=output-data-set
-
creates an output data set that contains polychoric correlation statistics. (Polychoric correlation between two observed binary
variables is also known as tetrachoric correlation.) This data set also includes means, standard deviations, and the number
of observations. The value of the _TYPE_ variable is 'CORR.'
-
OUTPLS=output-data-set
-
creates an output data set that contains polyserial correlation statistics. The contents of the output data set are similar
to those of the OUTPLC= data set.
-
OUTS=SAS-data-set
-
creates an output data set that contains Spearman correlation coefficients. The contents of the output data set are similar
to those of the OUTP= data set. When you specify the OUTS= option, the Spearman correlation coefficients will be displayed.
-
PEARSON
-
requests a table of Pearson product-moment correlations. The correlations range from –1 to 1. If you do not specify the HOEFFDING,
KENDALL, SPEARMAN, POLYCHORIC, POLYSERIAL, OUTH=, OUTK=, or OUTS= option, the CORR procedure produces Pearson product-moment
correlations by default. Otherwise, you must specify the PEARSON, ALPHA, COV, CSSCP, SSCP, or OUT= option for Pearson correlations.
Also, if a scatter plot or a scatter plot matrix is requested, the Pearson correlations will be displayed.
-
PLOTS <( MAXPOINTS=NONE | n )> = plot-request
PLOTS <( MAXPOINTS=NONE | n )> = ( plot-request <…plot-request> )
-
requests statistical graphics via the Output Delivery System (ODS).
ODS Graphics must be enabled before plots can be requested. For example:
ods graphics on;
proc corr data=Fitness plots=matrix(histogram);
run;
ods graphics off;
For more information about enabling and disabling ODS Graphics, see the section "Enabling and Disabling ODS Graphics" in
Chapter 21: Statistical Graphics Using ODS in SAS/STAT 13.2 User's Guide.
The global plot option MAXPOINTS= specifies that plots with elements that require processing more than n points be suppressed. The default is MAXPOINTS=5000. This limit is ignored if you specify MAXPOINTS=NONE. The plot request
options include the following:
-
ALL
-
produces all appropriate plots.
-
MATRIX <(matrix-options)>
-
requests a scatter plot matrix for variables. That is, the procedure displays a symmetric matrix plot with variables in the
VAR list if a WITH statement is not specified. Otherwise, the procedure displays a rectangular matrix plot with the WITH variables
appearing down the side and the VAR variables appearing across the top.
-
NONE
-
suppresses all plots.
-
SCATTER <(scatter-options)>
-
requests scatter plots for pairs of variables. That is, the procedure displays a scatter plot for each applicable pair of
distinct variables from the VAR list if a WITH statement is not specified. Otherwise, the procedure displays a scatter plot
for each applicable pair of variables, one from the WITH list and the other from the VAR list.
When a scatter plot or a scatter plot matrix is requested, the Pearson correlations will also be displayed.
The available matrix-options are as follows:
-
HIST | HISTOGRAM
-
displays histograms of variables in the VAR list (specified in the VAR statement) in the symmetric matrix plot.
-
NVAR=ALL | n
-
specifies the maximum number of variables in the VAR list to be displayed in the matrix plot, where n > 0. The NVAR=ALL option uses all variables in the VAR list. By default, NVAR=5.
-
NWITH=ALL | n
-
specifies the maximum number of variables in the WITH list (specified in the WITH statement) to be displayed in the matrix
plot, where n > 0. The NWITH=ALL option uses all variables in the WITH list. By default, NWITH=5.
If the resulting maximum number of variables in the VAR or WITH list is greater than 10, only the first 10 variables in the
list are displayed in the scatter plot matrix.
The available scatter-options are as follows:
-
ALPHA=

-
specifies the  values for the confidence or prediction ellipses to be displayed in the scatter plots, where
values for the confidence or prediction ellipses to be displayed in the scatter plots, where 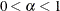 . For each
. For each  value specified, a (
value specified, a (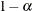 ) confidence or prediction ellipse is created. By default,
) confidence or prediction ellipse is created. By default, 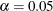 .
.
-
ELLIPSE=PREDICTION | CONFIDENCE | NONE
-
requests prediction ellipses for new observations (ELLIPSE=PREDICTION), confidence ellipses for the mean (ELLIPSE=CONFIDENCE),
or no ellipses (ELLIPSE=NONE) to be created in the scatter plots. By default, ELLIPSE=PREDICTION.
-
NOINSET
-
suppresses the default inset of summary information for the scatter plot. The inset table contains the number of observations
(Observations) and correlation.
-
NVAR=ALL | n
-
specifies the maximum number of variables in the VAR list (specified in the VAR statement) to be displayed in the plots, where
n > 0. The NVAR=ALL option uses all variables in the VAR list. By default, NVAR=5.
-
NWITH=ALL | n
-
specifies the maximum number of variables in the WITH list (specified in the WITH statement) to be displayed in the plots,
where n > 0. The NWITH=ALL option uses all variables in the WITH list. By default, NWITH=5.
If the resulting maximum number of variables in the VAR or WITH list is greater than 10, only the first 10 variables in the
list are displayed in the scatter plots.
-
POLYCHORIC <(options)>
-
requests a table of polychoric correlation coefficients. (Polychoric correlation between two observed binary variables is
also known as tetrachoric correlation.) A polychoric correlation measures the correlation between two unobserved, continuous variables that have a bivariate normal
distribution. Information about each unobserved variable is obtained through an observed ordinal variable that is derived
from the unobserved variable by classifying its values into a finite set of discrete, ordered values. If you specify a WEIGHT
statement, the POLYCHORIC option is not applicable.
You can specify the following options for computing polychoric correlation:
-
CONVERGE=p
-
specifies the convergence criterion. The value p must be between 0 and 1. The iterations are considered to have converged when the absolute change in the parameter estimates
between iteration steps is less than p for each parameter—that is, for the correlation and the thresholds for the unobserved continuous variable that define the
categories for the ordinal variable. By default, CONVERGE=0.0001.
-
MAXITER=number
-
specifies the maximum number of iterations. The iterations stop when the number of iterations exceeds number. By default, MAXITER=200.
-
NGROUPS=ALL | n
-
specifies the maximum number of groups allowed for each ordinal variable, where n > 1. NGROUPS=ALL allows an unlimited number of groups in each ordinal variable. Otherwise, if the number of groups exceeds
the specified number n, polychoric correlations are not computed for the affected pairs of variables. By default, NGROUPS=20.
-
POLYSERIAL <(options)>
-
requests a table of polyserial correlation coefficients. A polyserial correlation measures the correlation between two continuous variables with a bivariate normal distribution, where
one variable is observed and the other is unobserved. Information about the unobserved variable is obtained through an observed
ordinal variable that is derived from the unobserved variable by classifying its values into a finite set of discrete, ordered
values. If you specify a WEIGHT statement, the POLYSERIAL option is not applicable.
You can specify the following options for computing polyserial correlation:
-
CONVERGE=p
-
specifies the convergence criterion. The value p must be between 0 and 1. The iterations are considered to have converged when the absolute change in the parameter estimates
between iteration steps is less than p for each parameter—that is, for the correlation and the thresholds for the unobserved continuous variable that define the
categories for the ordinal variable. By default, CONVERGE=0.0001.
-
MAXITER=number
-
specifies the maximum number of iterations. The iterations stop when the number of iterations exceeds number. By default, MAXITER=200.
-
NGROUPS=ALL | n
-
specifies the maximum number of groups allowed for each ordinal variable, where n > 1. NGROUPS=ALL allows an unlimited number of groups in each ordinal variable. Otherwise, if the number of groups exceeds
the specified number n, polyserial correlations are not computed for the affected pairs of variables. By default, NGROUPS=20.
-
ORDINAL=WITH | VAR
-
specifies the ordinal variable list. The ORDINAL=WITH option specifies that the ordinal variables are provided in the WITH
statement, and the continuous variables are provided in the VAR statement. The ORDINAL=VAR option specifies that the ordinal
variables are provided in the VAR statement, and the continuous variables are provided in the WITH statement. By default,
ORDINAL=WITH.
-
RANK
-
displays the ordered correlation coefficients for each variable. Correlations are ordered from highest to lowest in absolute
value. If you specify the HOEFFDING option, the 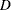 statistics are displayed in order from highest to lowest.
statistics are displayed in order from highest to lowest.
-
SINGULAR=p
-
specifies the criterion for determining the singularity of a variable if you use a PARTIAL statement. A variable is considered
singular if its corresponding diagonal element after Cholesky decomposition has a value less than p times the original unpartialled value of that variable. By default, SINGULAR=1E 8. The range of p is between 0 and 1.
8. The range of p is between 0 and 1.
-
SPEARMAN
-
requests a table of Spearman correlation coefficients based on the ranks of the variables. The correlations range from –1
to 1. If you specify a WEIGHT statement, the SPEARMAN option is invalid.
-
SSCP
-
displays a table of the sums of squares and crossproducts. When you specify the SSCP option, the Pearson correlations are
also displayed. If you specify the OUTP= option, the output data set contains a SSCP matrix and the corresponding _TYPE_ variable
value is 'SSCP.' If you use a PARTIAL statement, the unpartial SSCP matrix is displayed, and the output data set does not
contain an SSCP matrix.
-
VARDEF=DF | N | WDF | WEIGHT | WGT
-
specifies the variance divisor in the calculation of variances and covariances. The default is VARDEF=DF.
Table 2.2 displays available values and associated divisors for the VARDEF= option, where n is the number of nonmissing observations, k is the number of variables specified in the PARTIAL statement, and 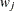 is the weight associated with the jth nonmissing observation.
is the weight associated with the jth nonmissing observation.
Table 2.2: Possible Values for the VARDEF= Option
|
Value
|
Description
|
Divisor
|
|
DF
|
Degrees of freedom
|
n - k - 1
|
|
N
|
Number of observations
|
n
|
|
WDF
|
Sum of weights minus one
|
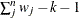
|
|
WEIGHT | WGT
|
Sum of weights
|
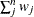
|

 Pearson Product-Moment CorrelationSpearman Rank-Order CorrelationKendall’s Tau-b Correlation CoefficientHoeffding Dependence CoefficientPartial CorrelationFisher’s z TransformationPolychoric CorrelationPolyserial CorrelationCronbach’s Coefficient AlphaConfidence and Prediction EllipsesMissing ValuesIn-Database ComputationOutput TablesOutput Data SetsODS Table NamesODS Graphics
Pearson Product-Moment CorrelationSpearman Rank-Order CorrelationKendall’s Tau-b Correlation CoefficientHoeffding Dependence CoefficientPartial CorrelationFisher’s z TransformationPolychoric CorrelationPolyserial CorrelationCronbach’s Coefficient AlphaConfidence and Prediction EllipsesMissing ValuesIn-Database ComputationOutput TablesOutput Data SetsODS Table NamesODS Graphics Computing Four Measures of AssociationComputing Correlations between Two Sets of VariablesAnalysis Using Fisher’s z TransformationApplications of Fisher’s z TransformationComputing Polyserial CorrelationsComputing Cronbach’s Coefficient AlphaSaving Correlations in an Output Data SetCreating Scatter PlotsComputing Partial Correlations
Computing Four Measures of AssociationComputing Correlations between Two Sets of VariablesAnalysis Using Fisher’s z TransformationApplications of Fisher’s z TransformationComputing Polyserial CorrelationsComputing Cronbach’s Coefficient AlphaSaving Correlations in an Output Data SetCreating Scatter PlotsComputing Partial Correlations