Example 12.5 Stackloss Data
The following example analyzes the three regressors of Brownlee (1965) stackloss data. By default, the MVE subroutine tries only 2000 randomly selected subsets in its search. There are, in total, 5985 subsets of 4 cases out of 21 cases. Here is the code:
title2 "***MVE for Stackloss Data***";
title3 "*** Use All Subsets***";
aa = { 1 80 27 89 42,
1 80 27 88 37,
1 75 25 90 37,
1 62 24 87 28,
1 62 22 87 18,
1 62 23 87 18,
1 62 24 93 19,
1 62 24 93 20,
1 58 23 87 15,
1 58 18 80 14,
1 58 18 89 14,
1 58 17 88 13,
1 58 18 82 11,
1 58 19 93 12,
1 50 18 89 8,
1 50 18 86 7,
1 50 19 72 8,
1 50 19 79 8,
1 50 20 80 9,
1 56 20 82 15,
1 70 20 91 15 };
a = aa[,2:4];
optn = j(9,1,.);
optn[1]= 2; /* ipri */
optn[2]= 1; /* pcov: print COV */
optn[3]= 1; /* pcor: print CORR */
optn[5]= -1; /* nrep: use all subsets */
call mve(sc,xmve,dist,optn,a);
Output 12.5.1 of the output shows the classical scatter and correlation matrix.
Output 12.5.1: Some Simple Statistics
| *** Brainlog Data: Do MCD, MVE *** |
| ***MVE for Stackloss Data*** |
| *** Use All Subsets*** |
| Classical Covariance Matrix | |||
|---|---|---|---|
| VAR1 | VAR2 | VAR3 | |
| VAR1 | 84.057142857 | 22.657142857 | 24.571428571 |
| VAR2 | 22.657142857 | 9.9904761905 | 6.6214285714 |
| VAR3 | 24.571428571 | 6.6214285714 | 28.714285714 |
| Classical Correlation Matrix | |||
|---|---|---|---|
| VAR1 | VAR2 | VAR3 | |
| VAR1 | 1 | 0.781852333 | 0.5001428749 |
| VAR2 | 0.781852333 | 1 | 0.3909395378 |
| VAR3 | 0.5001428749 | 0.3909395378 | 1 |
| Classical Mean | |
|---|---|
| VAR1 | 60.428571429 |
| VAR2 | 21.095238095 |
| VAR3 | 86.285714286 |
Output 12.5.2 shows the results of the optimization (complete subset sampling).
Output 12.5.2: Iteration History
| Subset | Singular | Best Criterion |
Percent |
|---|---|---|---|
| 1497 | 22 | 253.312431 | 25 |
| 2993 | 46 | 224.084073 | 50 |
| 4489 | 77 | 165.830053 | 75 |
| 5985 | 156 | 165.634363 | 100 |
| Observations of Best Subset | |||
|---|---|---|---|
| 7 | 10 | 14 | 20 |
| Initial MVE Location Estimates |
|
|---|---|
| VAR1 | 58.5 |
| VAR2 | 20.25 |
| VAR3 | 87 |
| Initial MVE Scatter Matrix | |||
|---|---|---|---|
| VAR1 | VAR2 | VAR3 | |
| VAR1 | 34.829014749 | 28.413143611 | 62.32560534 |
| VAR2 | 28.413143611 | 38.036950318 | 58.659393261 |
| VAR3 | 62.32560534 | 58.659393261 | 267.63348175 |
Output 12.5.3 shows the optimization results after local improvement.
Output 12.5.3: Table of MVE Results
| Robust MVE Location Estimates | |
|---|---|
| VAR1 | 56.705882353 |
| VAR2 | 20.235294118 |
| VAR3 | 85.529411765 |
| Robust MVE Scatter Matrix | |||
|---|---|---|---|
| VAR1 | VAR2 | VAR3 | |
| VAR1 | 23.470588235 | 7.5735294118 | 16.102941176 |
| VAR2 | 7.5735294118 | 6.3161764706 | 5.3676470588 |
| VAR3 | 16.102941176 | 5.3676470588 | 32.389705882 |
| Eigenvalues of Robust Scatter Matrix |
|
|---|---|
| VAR1 | 46.597431018 |
| VAR2 | 12.155938483 |
| VAR3 | 3.423101087 |
| Robust Correlation Matrix | |||
|---|---|---|---|
| VAR1 | VAR2 | VAR3 | |
| VAR1 | 1 | 0.6220269501 | 0.5840361335 |
| VAR2 | 0.6220269501 | 1 | 0.375278187 |
| VAR3 | 0.5840361335 | 0.375278187 | 1 |
Output 12.5.4 presents a table that contains the classical Mahalanobis distances, the robust distances, and the weights identifying the
outlying observations (that is, the leverage points when explaining ![]() with these three regressor variables).
with these three regressor variables).
Output 12.5.4: Mahalanobis and Robust Distances
| Classical Distances and Robust (Rousseeuw) Distances | |||
|---|---|---|---|
| Unsquared Mahalanobis Distance and | |||
| Unsquared Rousseeuw Distance of Each Observation | |||
| N | Mahalanobis Distances | Robust Distances | Weight |
| 1 | 2.253603 | 5.528395 | 0 |
| 2 | 2.324745 | 5.637357 | 0 |
| 3 | 1.593712 | 4.197235 | 0 |
| 4 | 1.271898 | 1.588734 | 1.000000 |
| 5 | 0.303357 | 1.189335 | 1.000000 |
| 6 | 0.772895 | 1.308038 | 1.000000 |
| 7 | 1.852661 | 1.715924 | 1.000000 |
| 8 | 1.852661 | 1.715924 | 1.000000 |
| 9 | 1.360622 | 1.226680 | 1.000000 |
| 10 | 1.745997 | 1.936256 | 1.000000 |
| 11 | 1.465702 | 1.493509 | 1.000000 |
| 12 | 1.841504 | 1.913079 | 1.000000 |
| 13 | 1.482649 | 1.659943 | 1.000000 |
| 14 | 1.778785 | 1.689210 | 1.000000 |
| 15 | 1.690241 | 2.230109 | 1.000000 |
| 16 | 1.291934 | 1.767582 | 1.000000 |
| 17 | 2.700016 | 2.431021 | 1.000000 |
| 18 | 1.503155 | 1.523316 | 1.000000 |
| 19 | 1.593221 | 1.710165 | 1.000000 |
| 20 | 0.807054 | 0.675124 | 1.000000 |
| 21 | 2.176761 | 3.657281 | 0 |
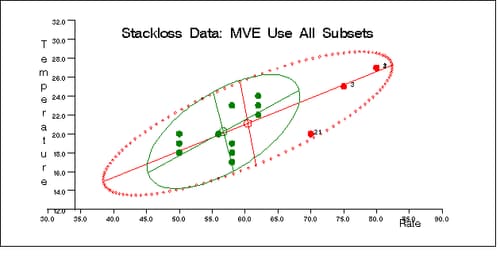
The following specification generates three bivariate plots of the classical and robust tolerance ellipsoids. They are shown in Output 12.5.5, Output 12.5.6, and Output 12.5.7, one plot for each pair of variables.
optn = j(9,1,.); optn[5]= -1;
vnam = { "Rate", "Temperature", "AcidConcent" };
filn = "stlmve";
titl = "Stackloss Data: Use All Subsets";
%include 'robustmc.sas';
call scatmve(2,optn,.9,a,vnam,titl,1,filn);
Output 12.5.5: Stackloss Data: Rate vs. Temperature (MVE)
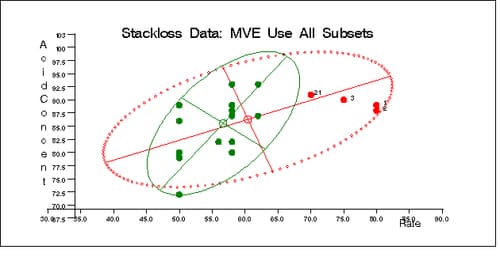
Output 12.5.6: Stackloss Data: Rate vs. Acid Concentration (MVE)
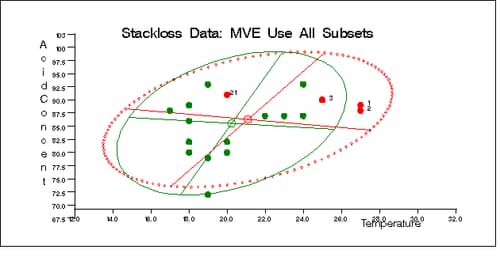
Output 12.5.7: Stackloss Data: Temperature vs. Acid Concentration (MVE)
You can also use the MCD method for the stackloss data as follows:
title2 "***MCD for Stackloss Data***"; title3 "*** Use 500 Random Subsets***"; a = aa[,2:4]; optn = j(8,1,.); optn[1]= 2; /* ipri */ optn[2]= 1; /* pcov: print COV */ optn[3]= 1; /* pcor: print CORR */ optn[5]= -1 ; /* nrep: use all subsets */ call mcd(sc,xmcd,dist,optn,a);
The optimization results are displayed in Output 12.5.8. The reweighted results are displayed in Output 12.5.9.
Output 12.5.8: MCD Results of Optimization
| *** Brainlog Data: Do MCD, MVE *** |
| ***MCD for Stackloss Data*** |
| *** Use 500 Random Subsets*** |
| 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 20 |
| MCD Location Estimate | ||
|---|---|---|
| VAR1 | VAR2 | VAR3 |
| 59.5 | 20.833333333 | 87.333333333 |
| MCD Scatter Matrix Estimate | |||
|---|---|---|---|
| VAR1 | VAR2 | VAR3 | |
| VAR1 | 5.1818181818 | 4.8181818182 | 4.7272727273 |
| VAR2 | 4.8181818182 | 7.6060606061 | 5.0606060606 |
| VAR3 | 4.7272727273 | 5.0606060606 | 19.151515152 |
| MCD Correlation Matrix | |||
|---|---|---|---|
| VAR1 | VAR2 | VAR3 | |
| VAR1 | 1 | 0.7674714142 | 0.4745347313 |
| VAR2 | 0.7674714142 | 1 | 0.4192963398 |
| VAR3 | 0.4745347313 | 0.4192963398 | 1 |
| Consistent Scatter Matrix | |||
|---|---|---|---|
| VAR1 | VAR2 | VAR3 | |
| VAR1 | 8.6578437815 | 8.0502757968 | 7.8983838007 |
| VAR2 | 8.0502757968 | 12.708297013 | 8.4553211199 |
| VAR3 | 7.8983838007 | 8.4553211199 | 31.998580526 |
Output 12.5.9: Final Reweighted MCD Results
| Reweighted Location Estimate | ||
|---|---|---|
| VAR1 | VAR2 | VAR3 |
| 59.5 | 20.833333333 | 87.333333333 |
| Reweighted Scatter Matrix | |||
|---|---|---|---|
| VAR1 | VAR2 | VAR3 | |
| VAR1 | 5.1818181818 | 4.8181818182 | 4.7272727273 |
| VAR2 | 4.8181818182 | 7.6060606061 | 5.0606060606 |
| VAR3 | 4.7272727273 | 5.0606060606 | 19.151515152 |
| Eigenvalues | ||
|---|---|---|
| VAR1 | VAR2 | VAR3 |
| 23.191069268 | 7.3520037086 | 1.3963209628 |
| Reweighted Correlation Matrix | |||
|---|---|---|---|
| VAR1 | VAR2 | VAR3 | |
| VAR1 | 1 | 0.7674714142 | 0.4745347313 |
| VAR2 | 0.7674714142 | 1 | 0.4192963398 |
| VAR3 | 0.4745347313 | 0.4192963398 | 1 |
The MCD robust distances and outlying diagnostic are displayed in Output 12.5.10. MCD identifies more leverage points than MVE.
Output 12.5.10: MCD Robust Distances
| Classical Distances and Robust (Rousseeuw) Distances | |||
|---|---|---|---|
| Unsquared Mahalanobis Distance and | |||
| Unsquared Rousseeuw Distance of Each Observation | |||
| N | Mahalanobis Distances | Robust Distances | Weight |
| 1 | 2.253603 | 12.173282 | 0 |
| 2 | 2.324745 | 12.255677 | 0 |
| 3 | 1.593712 | 9.263990 | 0 |
| 4 | 1.271898 | 1.401368 | 1.000000 |
| 5 | 0.303357 | 1.420020 | 1.000000 |
| 6 | 0.772895 | 1.291188 | 1.000000 |
| 7 | 1.852661 | 1.460370 | 1.000000 |
| 8 | 1.852661 | 1.460370 | 1.000000 |
| 9 | 1.360622 | 2.120590 | 1.000000 |
| 10 | 1.745997 | 1.809708 | 1.000000 |
| 11 | 1.465702 | 1.362278 | 1.000000 |
| 12 | 1.841504 | 1.667437 | 1.000000 |
| 13 | 1.482649 | 1.416724 | 1.000000 |
| 14 | 1.778785 | 1.988240 | 1.000000 |
| 15 | 1.690241 | 5.874858 | 0 |
| 16 | 1.291934 | 5.606157 | 0 |
| 17 | 2.700016 | 6.133319 | 0 |
| 18 | 1.503155 | 5.760432 | 0 |
| 19 | 1.593221 | 6.156248 | 0 |
| 20 | 0.807054 | 2.172300 | 1.000000 |
| 21 | 2.176761 | 7.622769 | 0 |
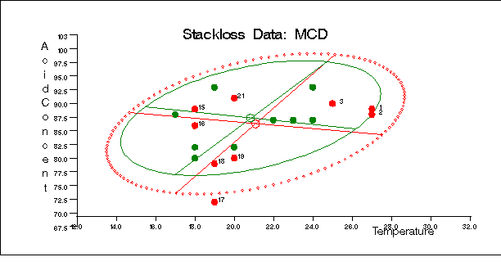
Similarly, you can use the SCATMCD routine to generate three bivariate plots of the classical and robust tolerance ellipsoids, one plot for each pair of variables. Here is the code:
optn = j(9,1,.); optn[5]= -1;
vnam = { "Rate", "Temperature", "AcidConcent" };
filn = "stlmcd";
titl = "Stackloss Data: Use All Subsets";
%include 'robustmc.sas';
call scatmcd(2,optn,.9,a,vnam,titl,1,filn);
Output 12.5.11, Output 12.5.12, and Output 12.5.13 display these plots.
Output 12.5.11: Stackloss Data: Rate vs. Temperature (MCD)
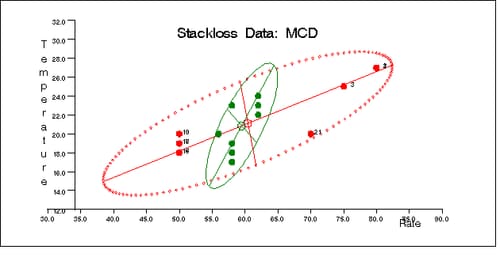
Output 12.5.12: Stackloss Data: Rate vs. Acid Concentration (MCD)
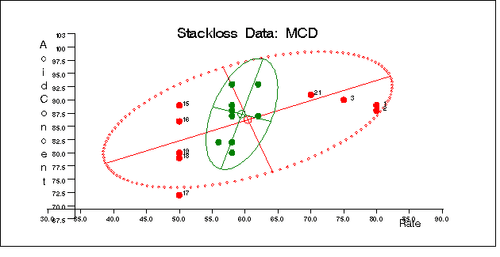
Output 12.5.13: Stackloss Data: Temperature vs. Acid Concentration (MCD)