The ENTROPY Procedure (Experimental)
- Overview
-
Getting Started

-
Syntax

-
Details
 Generalized Maximum Entropy Generalized Cross Entropy Moment Generalized Maximum Entropy Maximum Entropy-Based Seemingly Unrelated Regression Generalized Maximum Entropy for Multinomial Discrete Choice Models Censored or Truncated Dependent Variables Information Measures Parameter Covariance For GCE Parameter Covariance For GCE-M Statistical Tests Missing Values Input Data Sets Output Data Sets ODS Table Names ODS Graphics
Generalized Maximum Entropy Generalized Cross Entropy Moment Generalized Maximum Entropy Maximum Entropy-Based Seemingly Unrelated Regression Generalized Maximum Entropy for Multinomial Discrete Choice Models Censored or Truncated Dependent Variables Information Measures Parameter Covariance For GCE Parameter Covariance For GCE-M Statistical Tests Missing Values Input Data Sets Output Data Sets ODS Table Names ODS Graphics -
Examples

- References
| PROC ENTROPY Statement |
The following options can be specified in the PROC ENTROPY statement.
General Options
- COLLIN
requests that the collinearity diagnostics of the
 matrix be printed.
matrix be printed. - COVBEST=CROSS | GME | GMEM
specifies the method for producing the covariance matrix of parameters for output and for standard error calculations. GMEM and GME are aliases and are the default.
- GME | GCE
requests generalized maximum entropy or generalized cross entropy. This is the default estimation method.
- GMEM | GCEM
requests moment maximum entropy or the moment cross entropy.
- GMED
requests a variant of GME suitable for multinomial discrete choice models.
- MARKOV
- PURE
- SUR | ITSUR
specifies seemingly unrelated regression or iterated seemingly unrelated regression.
- VARDEF=N | WGT | DF | WDF
specifies the denominator to be used in computing variances and covariances. VARDEF=N specifies that the number of nonmissing observations be used. VARDEF=WGT specifies that the sum of the weights be used. VARDEF=DF specifies that the number of nonmissing observations minus the model degrees of freedom (number of parameters) be used. VARDEF=WDF specifies that the sum of the weights minus the model degrees of freedom be used. The default is VARDEF=DF.
Data Set Options
- DATA=SAS-data-set
specifies the input data set. Values for the variables in the model are read from this data set.
- PDATA=SAS-data-set
names the SAS data set that contains the data about priors and supports.
- OUT=SAS-data-set
names the SAS data set to contain the residuals from each estimation.
- OUTCOV
- COVOUT
writes the covariance matrix of the estimates to the OUTEST= data set in addition to the parameter estimates. The OUTCOV option is applicable only if the OUTEST= option is also specified.
- OUTEST=SAS-data-set
names the SAS data set to contain the parameter estimates and optionally the covariance of the estimates.
- OUTL=SAS-data-set
names the SAS data set to contain the estimated Lagrange multipliers for the models.
- OUTP=SAS-data-set
names the SAS data set to contain the support points and estimated probabilities.
- OUTS=SAS-data-set
names the SAS data set to contain the estimated covariance matrix of the equation errors. This is the covariance of the residuals computed from the parameter estimates.
- OUTSUSED=SAS-data-set
names the SAS data set to contain the
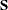 matrix used in the objective function definition. The OUTSUSED= data set is the same as the OUTS= data set for the methods that iterate the
matrix used in the objective function definition. The OUTSUSED= data set is the same as the OUTS= data set for the methods that iterate the 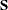 matrix.
matrix. - SDATA=SAS-data-set
specifies a data set that provides the covariance matrix of the equation errors. The matrix read from the SDATA= data set is used for the equation error covariance matrix (
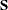 matrix) in the estimation. The SDATA= matrix is used to provide only the initial estimate of
matrix) in the estimation. The SDATA= matrix is used to provide only the initial estimate of 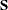 for the methods that iterate the S matrix.
for the methods that iterate the S matrix.
Printing Options
- ITPRINT
prints the parameter estimates, objective function value, and convergence criteria at each iteration.
- NOPRINT
suppresses the normal printed output but does not suppress error listings. Using any other print option turns the NOPRINT option off.
- PLOTS=global-plot-options | plot-request
-
controls the plots that the ENTROPY procedure produces. (For general information about ODS Graphics, see Chapter 21, Statistical Graphics Using ODS (SAS/STAT User's Guide).) The global-plot-options apply to all relevant plots generated by the ENTROPY procedure.
The global-plot-options supported by the ENTROPY procedure are as follows:
- ONLY
suppresses the default plots. Only the plots specifically requested are produced.
- UNPACKPANEL
breaks a graphic that is otherwise paneled into individual component plots.
The specific plot-request values supported by the ENTROPY procedure are as follows:
- ALL
requests that all plots appropriate for the particular analysis be produced. ALL is equivalent to specifying FITPLOT, COOKSD, QQ, RESIDUALHISTOGRAM, and STUDENTRESIDUAL.
- FITPLOT
plots the predicted and actual values.
- COOKSD
produces the Cook’s D plot.
produces a Q-Q plot of residuals.
- RESIDUALHISTOGRAM
plots the histogram of residuals.
- STUDENTRESIDUAL
plots the studentized residuals.
- NONE
suppresses all plots.
The default behavior is to plot all plots appropriate for the particular analysis (ALL) in a panel.
Options to Control the Minimization Process
The following options can be helpful if a convergence problem occurs for a given model and set of data. The ENTROPY procedure uses the nonlinear optimization subsystem (NLO) to perform the model optimizations. In addition to the options listed below, all options supported in the NLO subsystem can be specified on the ENTROPY procedure statement. See Chapter 6, Nonlinear Optimization Methods, for more details.
- CONVERGE=value
- GCONV=value
specifies the convergence criteria for S-iterated methods. The convergence measure computed during model estimation must be less than value before convergence is assumed. The default value is CONVERGE=0.001.
- DUAL | PRIMAL
specifies whether the optimization problem is solved using the dual or primal form. The dual form is the default.
- MAXITER=n
specifies the maximum number of iterations allowed. The default is MAXITER=100.
- MAXSUBITER=n
specifies the maximum number of subiterations allowed for an iteration. The MAXSUBITER= option limits the number of step halvings. The default is MAXSUBITER=30.
- METHOD=TR | NEWRAP | NRR | QN | CONGR | NSIMP | DBLDOG | LEVMAR
- TECHNIQUE=TR | NEWRAP | NRR | QN | CONGR | NSIMP | DBLDOG | LEVMAR
- TECH=TR | NEWRAP | NRR | QN | CONGR | NSIMP | DBLDOG | LEVMAR
specifies the iterative minimization method to use. METHOD=TR specifies the trust region method, METHOD=NEWRAP specifies the Newton-Raphson method, METHOD=NRR specifies the Newton-Raphson ridge method, and METHOD=QN specifies the quasi-Newton method. See Chapter 6, Nonlinear Optimization Methods, for more details about optimization methods. The default is METHOD=QN for the dual form and METHOD=NEWRAP for the primal form.
Note: This procedure is experimental.