The HPCDM Procedure (Experimental)
PROC HPCDM options ;
The PROC HPCDM statement invokes the procedure. You can specify the following options, which are listed in alphabetical order.
-
ADJUSTEDSEVERITY=symbol-name
ADJSEV=symbol-name -
names the symbol that represents the adjusted severity value in the SAS programming statements that you specify. The symbol-name is a SAS name that conforms to the naming conventions of a SAS variable. For more information, see the section Programming Statements.
- COUNTSTORE=SAS-item-store
-
names the item store that contains all the information about the frequency (count) model. The COUNTREG procedure generates this item store when you use the STORE statement.
The exogenous variables in the frequency model, if any, are deduced from this item store. The DATA= data set must contain all those variables.
You must specify this option if you do not specify the EXTERNALCOUNTS statement. This option is ignored if you specify the EXTERNALCOUNTS statement, because PROC HPCDM does not need to simulate frequency counts internally when you specify externally simulated counts.
In SAS/ETS 13.1, PROC COUNTREG can create an item store only when you fit one count model and when you do not specify the BY statement in PROC COUNTREG. So if you specify the COUNTSTORE= option, then you cannot specify the BY statement in PROC HPCDM, and vice versa.
- DATA=SAS-data-set
-
names the input data set that contains the values of regression variables in frequency or severity models and severity adjustment variables that you use in the programming statements.
The DATA= data set specifies information about the scenario for which you want to estimate the aggregate loss distribution. The contents of the data set are interpreted differently based on whether you specify the EXTERNALCOUNTS statement. For more information, see the section Specifying Scenario Data in the DATA= Data Set.
- NOPRINT
-
turns off all displayed and graphical output. If you specify this option, then PROC HPCDM ignores any value that you specify for the PRINT= or PLOTS= option.
-
NPERTURBEDSAMPLES=number
NPERTURB=number -
requests that parameter perturbation analysis be conducted. The model parameters are perturbed the specified number of times and a separate full sample is simulated for each set of perturbed parameter values. The summary statistics and percentiles are computed for each such perturbed sample, and their values are aggregated across the samples to compute the mean and standard deviation of each summary statistic and percentile.
The parameter perturbation procedure makes random draws of parameter values from a multivariate normal distribution if the covariance estimates of the parameters are available in the SEVERITYEST= data set for the severity model and in the COUNTSTORE= store for the count model. If covariance estimates are not available, then for each parameter, a random draw is made from the univariate normal distribution that has mean and standard deviation equal to the point estimate and the standard error, respectively, of that parameter. If neither covariance nor standard error estimates are available, then perturbation analysis is not conducted.
If you specify the PRINT=ALL or PRINT=PERTURBSUMMARY option, then the summary of perturbation analysis is printed for the core summary statistics and the percentiles of the aggregate loss distribution. If you specify the OUTSUM statement, then the requested summary statistics are written to the OUTSUM= data set for each perturbed sample. You can also optionally request that each perturbed sample be written in its entirety to the OUT= data set by specifying the PERTURBOUT option in the OUTPUT statement.
For more information on the parameter perturbation analysis, see the section Parameter Perturbation Analysis.
-
NREPLICATES=number
NREP=number -
specifies a number that controls the size of the compound distribution sample that PROC HPCDM simulates. The number is interpreted differently based on whether you specify the EXTERNALCOUNTS statement.
If you do not specify the EXTERNALCOUNTS statement, then the sample size is equal to the number that you specify for this option. If you do not specify this option, then a default value of 100,000 is used.
If you specify the EXTERNALCOUNTS statement, then the number of replicates that you specify in the DATA= data set is multiplied by the number that you specify for this option to get the total size of the compound distribution sample. If you do not specify this option, then a default value of 1 is used.
- PCTLDEF=percentile-method
-
specifies the method to compute the percentiles of the compound distribution. The percentile-method can be 1, 2, 3, 4, or 5. The default method is 5. For more information, see the description of the PCTLDEF= option in the UNIVARIATE procedure in the Base SAS Procedures Guide: Statistical Procedures.
-
PLOTS <(global-plot-options)> =plot-request-option
PLOTS <(global-plot-options)> =(plot-request-option …plot-request-option) -
specifies the desired graphical output.
By default, the HPCDM procedure produces no graphical output.
You can specify the following global-plot-option:
If you specify more than one plot-request-option, then separate them with spaces and enclose them in parentheses. The following plot-request-options are available:
- ALL
-
displays all the graphical output.
-
CONDITIONALDENSITY (conditional-density-plot-options)
CONDPDF (conditional-density-plot-options) -
prepares a group of plots of the conditional density functions estimates. The group contains at most three plots, each conditional on the value of the aggregate loss being in one of the three regions that are defined by the quantiles that you specify in the following conditional-density-plot-options:
- LEFTQ=number
-
specifies the quantile in the range (0,1) that marks the end of the left-tail region. If you specify a value of
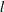 for number, then the left-tail region is defined as the set of values that are less than or equal to
for number, then the left-tail region is defined as the set of values that are less than or equal to 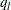 , where
, where 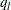 is the
is the 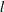 th quantile. For the left-tail region, nonparametric estimates of the conditional probability density function
th quantile. For the left-tail region, nonparametric estimates of the conditional probability density function ![$f^ l_ S(s) = \Pr [S=s|S \leq q_ l]$](images/etshpug_hpcdm0024.png) are plotted. The value of
are plotted. The value of 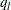 is estimated by the
is estimated by the 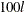 th percentile of the simulated compound distribution sample.
th percentile of the simulated compound distribution sample.
If you do not specify this option or you specify a missing value for this option, then the left-tail region is not plotted.
- RIGHTQ=number
-
specifies the quantile in the range (0,1) that marks the beginning of the right-tail region. If you specify a value of
 for number, then the right-tail region is defined as the set of values that are greater than
for number, then the right-tail region is defined as the set of values that are greater than 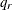 , where
, where 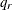 is the
is the  th quantile. For the right-tail region, nonparametric estimates of the conditional probability density function
th quantile. For the right-tail region, nonparametric estimates of the conditional probability density function ![$f^ r_ S(s) = \Pr [S=s|S > q_ r]$](images/etshpug_hpcdm0028.png) are plotted. The value of
are plotted. The value of 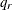 is estimated by the
is estimated by the 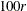 th percentile of the simulated compound distribution sample.
th percentile of the simulated compound distribution sample.
If you do not specify this option or you specify a missing value for this option, then the right-tail region is not plotted.
You must specify nonmissing value for at least one of the preceding two options. For the region between the LEFTQ= and RIGHTQ= quantiles, which is referred to as the central or body region, nonparametric estimates of the conditional probability density function
![$f^ c_ S(s) = \Pr [S=s|q_ l < S \leq q_ r]$](images/etshpug_hpcdm0030.png) are plotted. If you do not specify a LEFTQ= value, then
are plotted. If you do not specify a LEFTQ= value, then 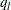 is assumed to be 0. If you do not specify a RIGHTQ= value, then
is assumed to be 0. If you do not specify a RIGHTQ= value, then 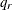 is assumed to be
is assumed to be 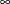 .
.
- DENSITY
-
prepares a plot of the nonparametric estimates of the probability density function (in particular, histogram and kernel density estimates) of the compound distribution.
- EDF <(edf-plot-option)>
-
prepares a plot of the nonparametric estimates of the cumulative distribution function of the compound distribution.
You can request that the confidence interval be plotted by specifying the following edf-plot-option:
- NONE
-
displays none of the graphical output. If you specify this option, then it overrides all other plot request options. The default graphical output is also suppressed.
Note that if the simulated sample size is large, then it can take a significant amount of time and memory to prepare the plots.
-
PRINT <(global-display-option)> =display-option
PRINT <(global-display-option)> =(display-option …display-option) -
specifies the desired displayed output. If you specify more than one display-option, then separate them with spaces and enclose them in parentheses.
You can specify the following global-display-option:
You can specify the following display-options:
- ALL
-
displays all the output.
- NONE
-
displays none of the output. If you specify this option, then it overrides all other display options. The default displayed output is also suppressed.
- PERCENTILES
-
displays the percentiles of the compound distribution sample. This includes all the predefined percentiles, percentiles that you request in the OUTSUM statement, and percentiles that you specify for preparing conditional density plots.
- PERTURBSUMMARY
-
displays the mean and standard deviation of the summary statistics and percentiles that are taken across all the samples produced by perturbing the model parameters. This option is valid only if you specify the NPERTURBEDSAMPLES= option in the PROC HPCDM statement.
- SUMMARYSTATISTICS | SUMSTAT
-
displays the summary statistics of the compound distribution sample.
If you do not specify the PRINT= option or the ONLY global-display-option, then the default displayed output is equivalent to specifying PRINT=(SUMMARYSTATISTICS).
- SEED=number
-
specifies the integer to use as the seed in generating the pseudo-random numbers that are used for simulating severity and frequency values. If you do not specify the seed or if number is negative or 0, then the time of day from the computer’s clock is used as the seed.
- SEVERITYEST=SAS-data-set
-
names the input data set that contains the parameter estimates for the severity model. The format of this data set must be the same as the OUTEST= data set that is produced by the SEVERITY procedure.
The names of the regression variables in the scale regression model, if any, are deduced from this data set. In particular, PROC HPCDM assumes that all the variables in the SEVERITYEST= data set that do not appear in the following list are scale regression variables:
-
BY variables
-
_MODEL_, _TYPE_, _NAME_, and _STATUS_ variables
-
variables that represent distribution parameters
The DATA= data set must contain all the regressors in the scale regression model.
To ensure that PROC HPCDM correctly matches the values of regressors and the values of regression parameter estimates, you might need to rename the regressors in the DATA= data set so that their names match the names of the regressors that you specify in the SCALEMODEL statement of the PROC SEVERITY step that fits the severity model.
-
- VARDEF=divisor
-
specifies the divisor to use in the calculation of variance, standard deviation, kurtosis, and skewness of the compound distribution sample. If the sample size is
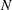 , then you can specify one of the following values for the divisor:
, then you can specify one of the following values for the divisor:
For more information, see the section Descriptive Statistics.
