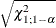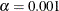The ROBUSTREG Procedure
MODEL Statement
-
<label:> MODEL response = <effects> </ options>;
You can specify main effects and interaction terms in the MODEL statement, as in the GLM procedure (see Chapter 46: The GLM Procedure).
The optional label, which must be a valid SAS name, is used to label the model in the OUTEST data set.
Table 98.9 summarizes the options available in the MODEL statement.
Table 98.9: MODEL Statement Options
|
Option |
Description |
|---|---|
|
Specifies the significance level |
|
|
Produces the estimated correlation matrix |
|
|
Produces the estimated covariance matrix |
|
|
Specifies the multiplier of the cutoff value for outlier detection |
|
|
Requests the outlier diagnostics |
|
|
Specifies the failure-ratio threshold |
|
|
Displays the iteration history |
|
|
Requests an analysis of leverage points |
|
|
Suppresses the computation of goodness-of-fit statistics |
|
|
Specifies no-intercept regression |
|
|
Specifies the tolerance for testing singularity |
You can specify the following options for the model fit.


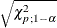
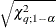
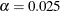
![$[(3n+p+1)/4]$](images/statug_rreg0017.png)