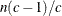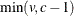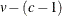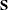The HPCANDISC Procedure
PROC HPCANDISC Statement
-
PROC HPCANDISC <options>;
The PROC HPCANDISC statement invokes the HPCANDISC procedure. Optionally, it also identifies input and output data sets, specifies the analyses performed, and controls displayed output. Table 50.1 summarizes the options available in the PROC HPCANDISC statement.
Table 50.1: PROC HPCANDISC Statement Options
|
Option |
Description |
|---|---|
|
Specify Data Sets |
|
|
Specifies the input data set |
|
|
Specifies the output data set that contains canonical scores |
|
|
Specifies the output statistics data set |
|
|
Specify Details of Analysis |
|
|
Specifies the number of canonical variables |
|
|
Specifies a prefix for naming the canonical variables |
|
|
Specifies the singularity criterion |
|
|
Control Displayed Output |
|
|
Displays all output |
|
|
Displays univariate statistics |
|
|
Displays between correlations |
|
|
Displays between covariances |
|
|
Displays between SSCPs |
|
|
Displays squared Mahalanobis distances |
|
|
Suppresses all displayed output |
|
|
Displays pooled correlations |
|
|
Displays pooled covariances |
|
|
Displays pooled SSCPs |
|
|
Suppresses some displayed output |
|
|
Displays simple descriptive statistics |
|
|
Displays standardized class means |
|
|
Displays total correlations |
|
|
Displays total covariances |
|
|
Displays total SSCPs |
|
|
Displays within correlations |
|
|
Displays within covariances |
|
|
Displays within SSCPs |
|
The following list provides details about these options.