The MIXED Procedure
PARMS Statement
PARMS (value-list)…</ options> ;
The PARMS statement specifies initial values for the covariance parameters, or it requests a grid search over several values of these parameters. You must specify the values in the order in which they appear in the “Covariance Parameter Estimates” table.
The value-list specification can take any of several forms:
- m
-
a single value
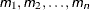
-
several values
- m to n
-
a sequence where m equals the starting value, n equals the ending value, and the increment equals 1
- m to n by i
-
a sequence where m equals the starting value, n equals the ending value, and the increment equals i
 to
to 
-
mixed values and sequences
You can use the PARMS statement to input known parameters. Referring to the split-plot example ( Example 59.1), suppose the three variance components are known to be 60, 20, and 6. The SAS statements to fix the variance components at these values are as follows:
proc mixed data=sp noprofile; class Block A B; model Y = A B A*B; random Block A*Block; parms (60) (20) (6) / noiter; run;
The NOPROFILE option requests PROC MIXED to refrain from profiling the residual variance parameter during its calculations, thereby enabling
its value to be held at 6 as specified in the PARMS statement. The NOITER option prevents any Newton-Raphson iterations so that the subsequent results are based on the given variance components.
You can also specify known parameters of ![]() by using the GDATA= option in the RANDOM statement.
by using the GDATA= option in the RANDOM statement.
If you specify more than one set of initial values, PROC MIXED performs a grid search of the likelihood surface and uses the best point on the grid for subsequent analysis. Specifying a large number of grid points can result in long computing times. The grid search feature is also useful for exploring the likelihood surface. (See Example 59.3.)
The results from the PARMS statement are the values of the parameters on the specified grid (denoted by CovP1–CovPn), the residual variance (possibly estimated) for models with a residual variance parameter, and various functions of the likelihood.
The ODS name of the “Parameter Search” table is ParmSearch.
Table 59.13 summarizes the options available in the PARMS statement.
Table 59.13: PARMS Statement Options
|
Option |
Description |
|---|---|
|
Holds parameter values equal to the specified values |
|
|
Evaluates the log determinant of the Hessian matrix |
|
|
Specifies lower boundary constraints |
|
|
Removes boundary constraints on covariance parameters |
|
|
Performs inferences using the best value from the grid search |
|
|
Suppresses the “Parameter Search” table |
|
|
Specifies a different computational method for the residual variance |
|
|
Requests starting values corresponding to the usual general linear model |
|
|
Reads in covariance parameter values from a SAS data set |
|
|
Indicates that ratios with the residual variance are specified |
|
|
Specifies upper boundary constraints |
You can specify the following options in the PARMS statement after a slash (/).
