| The GLMSELECT Procedure |
| PROC GLMSELECT Statement |
Table 42.1 lists the options available in the PROC GLMSELECT statement.
Option |
Description |
|---|---|
Data Set Options |
|
Names a data set to use for the regression |
|
Sets the maximum number of macro variables produced |
|
Names a data set that contains test data |
|
Names a data set that contains validation data |
|
ODS Graphics Options |
|
Produces ODS graphical displays |
|
Other Options |
|
Requests a data set that contains the design matrix |
|
Sets the length of effect names in tables and output data sets |
|
Suppresses displayed output including plots |
|
Sets the seed used for pseudo-random number generation |
|
Following are explanations of the options that you can specify in the PROC GLMSELECT statement (in alphabetical order).
- DATA=SAS-data-set
names the SAS data set to be used by PROC GLMSELECT. If the DATA= option is not specified, PROC GLMSELECT uses the most recently created SAS data set. If the named data set contains a variable named _ROLE_, then this variable is used to assign observations for training, validation, and testing roles. See the section Using Validation and Test Data for details on using the _ROLE_ variable.
- MAXMACRO=n
specifies the maximum number of macro variables with selected effects to create. By default, MAXMACRO=100. PROC GLMSELECT saves the list of selected effects in a macro variable, &_GLSIND. Say your input effect list consists of x1-x10. Then &_GLSIND would be set to x1 x3 x4 x10 if, for example, the first, third, fourth, and tenth effects were selected for the model. This list can be used, for example, in the model statement of a subsequent procedure. If you specify the OUTDESIGN= option in the PROC GLMSELECT statement, then PROC GLMSELECT saves the list of columns in the design matrix in a macro variable named &_GLSMOD.
With BY processing, one macro variable is created for each BY group, and the macro variables are indexed by the BY group number. The MAXMACRO= option can be used to either limit or increase the number of these macro variables when you are processing data sets with many BY groups.
With no BY processing, PROC GLMSELECT creates the following:
_GLSIND
selected effects
_GLSIND1
selected effects
_GLSMOD
design matrix columns
_GLSMOD1
design matrix columns
_GLSNUMBYS
number of BY groups
_GLSNUMMACROBYS
number of _GLSIND
 macro variables actually made
macro variables actually made With BY processing, PROC GLMSELECT creates the following:
_GLSIND
selected effects for BY group 1
_GLSIND1
selected effects for BY group 1
_GLSIND2
selected effects for BY group 2
.
.
.
_GLSIND

selected effects for BY group
 , where a number is substituted for
, where a number is substituted for 
_GLSMOD
design matrix columns for BY group 1
_GLSMOD1
design matrix columns for BY group 1
_GLSMOD2
design matrix columns for BY group 2
.
.
.
_GLSMOD

design matrix columns for BY group
 , where a number is substituted for
, where a number is substituted for 
_GLSNUMBYS
 , the number of BY groups
, the number of BY groups _GLSNUMMACROBYS
the number
 of _GLSIND
of _GLSIND macro variables actually made. This value can be less than _GLSNUMBYS =
macro variables actually made. This value can be less than _GLSNUMBYS =  , and it is less than or equal to the MAXMACRO= value.
, and it is less than or equal to the MAXMACRO= value.See the section Macro Variables Containing Selected Models for further details.
- NOPRINT
- NAMELEN=n
specifies the length of effect names in tables and output data sets to be n characters long, where n is a value between 20 and 200 characters. The default length is 20 characters.
- OUTDESIGN <(options)><=SAS-data-set>
creates a data set that contains the design matrix. By default, the GLMSELECT procedure includes in the OUTDESIGN data set the
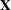 matrix corresponding to the parameters in the selected model. Two schemes for naming the columns of the design matrix are available. In the first scheme, names of the parameters are constructed from the parameter labels that appear in the "ParameterEstimates" table. This naming scheme is the default when you do not request BY processing and is not available when you do use BY processing. In the second scheme, the design matrix column names consist of a prefix followed by an index. The default naming prefix is "_X". You can specify the following options in parentheses to control the contents of the OUTDESIGN data set:
matrix corresponding to the parameters in the selected model. Two schemes for naming the columns of the design matrix are available. In the first scheme, names of the parameters are constructed from the parameter labels that appear in the "ParameterEstimates" table. This naming scheme is the default when you do not request BY processing and is not available when you do use BY processing. In the second scheme, the design matrix column names consist of a prefix followed by an index. The default naming prefix is "_X". You can specify the following options in parentheses to control the contents of the OUTDESIGN data set:- ADDINPUTVARS
requests that all variables in the input data set be included in the OUTDESIGN= data set.
- FULLMODEL
specifies that parameters corresponding to all the effects specified in the MODEL statement be included in the OUTDESIGN= data set. By default, only parameters corresponding to the selected model are included.
- NAMES
produces a table associating columns in the OUTDESIGN data set with the labels of the parameters they represent.
- PREFIX<=prefix>
requests that the design matrix column names consist of a prefix followed by an index. The default naming prefix is "_X". You can optionally specify a different prefix.
- PARMLABELSTYLE=options
specifies how parameter names and labels are constructed for nested and crossed effects.
The following options are available:
- INTERLACED <(SEPARATOR=quoted string)>
forms parameter names and labels by positioning levels of classification variables and constructed effects adjacent to the associated variable or constructed effect name and using " * " as the delimiter for both crossed and nested effects. This style of naming parameters and labels is used in the TRANSREG procedure. You can request truncation of the classification variable names used in forming the parameter names and labels by using the CPREFIX= and LPREFIX= options in the CLASS statement. You can use the SEPARATOR= suboption to change the delimiter between the crossed variables in the effect. PARMLABELSTYLE=INTERLACED is not supported if you specify the SPLIT option in an EFFECT statement or a CLASS statement. The following are examples of the parameter labels in this style (Age is a continuous variable, Gender and City are classification variables):
Age Gender male * City Beijing City London * Age
- SEPARATE
specifies that in forming parameter names and labels, the effect name appears before the levels associated with the classification variables and constructed effects in the effect. You can control the length of the effect name by using the NAMELEN= option in the PROC GLMSELECT statement. In forming parameter labels, the first level that is displayed is positioned so that it starts at the same offset in every parameter label—this enables you to easily distinguish the effect name from the levels when the parameter labels are displayed in a column in the "Parameter Estimates" table. This style of labeling is used in the GLM procedure and is the default if you do not specify the PARMLABELSTYLE option. The following are examples of the parameter labels in this style (Age is a continuous variable, Gender and City are classification variables):
Age Gender*City male Beijing Age*City London
- SEPARATECOMPACT
requests the same parameter naming and labeling scheme as PARMLABELSTYLE=SEPARATE except that the first level in the parameter label is separated from the effect name by a single blank. This style of labeling is used in the PLS procedure. The following are examples of the parameter labels in this style (Age is a continuous variable, Gender and City are classification variables):
Age Gender*City male Beijing Age*City London
- PLOTS <(global-plot-options)> <= plot-request <(options)>>
- PLOTS <(global-plot-options)> <= (plot-request <(options)> <... plot-request <(options)>>)>
controls the plots produced through ODS Graphics. When you specify only one plot request, you can omit the parentheses around the plot request. Here are some examples:
plots=all plots=coefficients(unpack) plots(unpack)=(criteria candidates)
You must enable ODS Graphics before requesting plots as shown in the following example. For general information about ODS Graphics, see Chapter 21, Statistical Graphics Using ODS.
ods graphics on; proc glmselect plots=all; model y = x1-x100; run; ods graphics off;
Global Plot OptionsThe global-options apply to all plots generated by the GLMSELECT procedure, unless it is altered by a specific-plot-option.
- ENDSTEP=n
specifies that the step ranges shown on the horizontal axes of plots terminates at specified step. By default, the step range shown terminates at the final step of the selection process. If you specify the ENDSTEP= option as both a global plot option and a specific plot option, then the ENDSTEP= value on the specific plot is used.
- LOGP | LOGPVALUE
requests that the natural logarithm of the entry and removal significance levels be displayed. This option is ignored if the select criterion is not significance level.
- MAXSTEPLABEL=n
specifies the maximum number of characters beyond which labels of effects on plots are truncated.
- MAXPARMLABEL= n
specifies the maximum number of characters beyond which parameter labels on plots are truncated.
- STARTSTEP=n
specifies that the step ranges shown on the horizontal axes of plots start at the specified step. By default, the step range shown starts at the initial step of the selection process. If you specify the STARTSTEP= option both as a global plot option and a specific plot option, then the STARTSTEP= value on the specific plot is used.
- STEPAXIS=EFFECT | NORMB | NUMBER
specifies the horizontal axis to be used on the plots, where this axis represents the sequence of entering or departing effects.
- STEPAXIS=EFFECT
requests that each step be labeled by a prefix followed by the name of the effect that enters or leaves at that step. The prefix consists of the step number followed by a "+" sign or a "-" sign depending on whether the effect enters or leaves at that step.
- STEPAXIS=NORMB
is valid only with LAR and LASSO selection methods and requests that the horizontal axis value at step
 be the L1 norm of the parameters at step
be the L1 norm of the parameters at step  , normalized by the L1 norm of the parameters at the final step.
, normalized by the L1 norm of the parameters at the final step. - STEPAXIS=NUMBER
requests that each step be labeled by the step number.
- UNPACK
suppresses paneling. By default, multiple plots can appear in some output panels. Specify UNPACK to get each plot individually. You can also specify UNPACK as a suboption with CRITERIA and COEFFICIENTS.
Specific Plot OptionsThe following listing describes the specific plots and their options.
- ALL
requests that all default plots be produced. Note that candidate plots are produced only if you specify DETAILS=STEPS or DETAILS=ALL in the MODEL statement.
- ASE | ASEPLOT <(aseplot-option)>
plots the progression of the average square error on the training data, and the test and validation data whenever these data are provided with the TESTDATA= and VALDATA= options or are produced by using a PARTITION statement. The following aseplot-option option is available:
- STEPAXIS=EFFECT | NORMB | NUMBER
specifies the horizontal axis to be used.
- CANDIDATES | CANDIDATESPLOT <(candidatesplot-options)>
produces a needle plot of the select criterion values for the candidates for entry or removal at each step of the selection process, ordered from best to worst. Candidates plots are not available if you specify SELECTION=NONE, SELECTION=LAR, or SELECTION=LASSO in the MODEL statement, or if you have not specified DETAILS=ALL or DETAILS=STEPS in the MODEL statement. The following candidatesplot-options are available:
- LOGP | LOGPVALUE
requests that the natural logarithm of the entry and removal significance levels be displayed. This option is ignored if the select criterion is not significance level.
- SHOW=number
specifies the maximum number of candidates displayed at each step. The default is SHOW=10.
- COEFFICIENTS | COEFFICIENTPANEL <(coefficientPanel-options)>
plots a panel of two plots. The upper plot shows the progression of the parameter values as the selection process proceeds. The lower plot shows the progression of the CHOOSE= criterion. If no choose criterion is in effect, then the AICC criterion is displayed. The following coefficientPanel-options are available:
- LABELGAP=percentage
specifies the percentage of the vertical axis range that forms the minimum gap between successive parameter labels at the final step of the coefficient progression plot. If the values of more than one parameter at the final step are closer than this gap, then the labels on all but one of these parameters is suppressed. The default value is LABELGAP=5. Planned enhancements to the automatic label collision avoidance algorithm will obviate the need for this option in future releases of the GLMSELECT procedure.
- LOGP | LOGPVALUE
requests that the natural logarithm of the entry and removal significance levels be displayed if the choose criterion is significance level.
- STEPAXIS=EFFECT | NORMB | NUMBER
specifies the horizontal axis to be used.
- UNPACK | UNPACKPANEL
displays the coefficient progression and the choose criterion progression in separate plots.
- CRITERIA | CRITERIONPANEL <(criterionPanel-options)>
plots a panel of model fit criteria. The criteria that are displayed are ADJRSQ, AIC, AICC, and SBC, as well as any other criteria that are named in the CHOOSE=, SELECT=, STOP=, or STATS= option in the MODEL statement. The following criterionPanel-options are available:
- STEPAXIS=EFFECT | NORMB | NUMBER
specifies the horizontal axis to be used.
- UNPACK | UNPACKPANEL
displays each criterion progression on a separate plot.
- EFFECTSELECTPCT <(effectSelectPct-options)>
requests a bar chart whose bars correspond to effects that are selected in at least one sample when you use the MODELAVERAGE statement. The length of a bar corresponds to the percentage of samples where the selected model contains the effect the bar represents. The EFFECTSELECTPCT option is ignored if you do not specify a MODELAVERAGE statement. The following effectSelectPct-options are available:
- MINPCT=percent
specifies that effects that appear in fewer than the specified percentage of the sample selected models not be included in the plot. By default, effects that are shown in the "EffectSelectPct" table are displayed.
- ORDER=ASCENDING | DESCENDING | MODEL
specifies the ordering of the effects in the bar chart. ORDER=MODEL specifies that effects appear in the order in which they appear in the MODEL statement. ORDER=ASCENDING | DESCENDING specifies that the effects are shown in ascending or descending order of the number of samples in which the effects appear in the selected model. The default is ORDER=DESCENDING.
- NONE
suppresses all plots.
- PARMDIST <(parmDist-options)>
produces a panel that shows histograms and box plots of the parameter estimate values across samples when you use a MODELAVERAGE statement. There is a histogram and box plot for each parameter that appears in the "AvgParmEst" table. The PARMDIST option is ignored if you do not specify a MODELAVERAGE statement. The following parmDist-options are available:
- MINPCT=percent
specifies that distributions be shown only for parameters whose estimates are nonzero in at least the specified percentage of the selected models. By default, distributions are shown for the all parameters that appear in the "AvgParmEst" table.
- ORDER=ASCENDING | DESCENDING | MODEL
specifies the ordering of the parameters in the panels. ORDER=MODEL specifies that parameters be shown in the order in which the corresponding effects appear in the MODEL statement. ORDER=ASCENDING | DESCENDING specifies that the parameters be shown in an ascending or descending order of the number of samples in which the parameter estimate is nonzero. The default is ORDER=DESCENDING.
- NOBOXPLOTS
suppress the box plots.
- PLOTSPERPANEL=number
specifies the maximum number of parameter distributions that appear in a panel. If the number of relevant parameters is greater than number, then multiple panels are produced. Valid values are 1–16 with 9 as the default.
- UNPACK
specifies that the distribution for each relevant parameter be shown in a separate plot.
- SEED=number
specifies an integer used to start the pseudo-random number generator for resampling the data, random cross validation, and random partitioning of data for training, testing, and validation. If you do not specify a seed, or if you specify a value less than or equal to zero, the seed is generated from reading the time of day from the computer’s clock.
- TESTDATA=SAS-data-set
names a SAS data set containing test data. This data set must contain all the variables specified in the MODEL statement. Furthermore, when a BY statement is used and the TESTDATA=data set contains any of the BY variables, then the TESTDATA= data set must also contain all the BY variables sorted in the order of the BY variables. In this case, only the test data for a specific BY group is used with the corresponding BY group in the analysis data. If the TESTDATA= data set contains none of the BY variables, then the entire TESTDATA = data set is used with each BY group of the analysis data.
If you specify a TESTDATA=data set, then you cannot also reserve observations for testing by using a PARTITION statement.
- VALDATA=SAS-data-set
names a SAS data set containing validation data. This data set must contain all the variables specified in the MODEL statement. Furthermore, when a BY statement is used and the VALDATA=data set contains any of the BY variables, then the VALDATA= data set must also contain all the BY variables sorted in the order of the BY variables. In this case, only the validation data for a specific BY group are used with the corresponding BY group in the analysis data. If the VALDATA= data set contains none of the BY variables, then the entire VALDATA = data set is used with each BY group of the analysis data.
If you specify a VALDATA=data set, then you cannot also reserve observations for validation by using a PARTITION statement.
Copyright © SAS Institute, Inc. All Rights Reserved.