| The MI Procedure |
| Producing Monotone Missingness with the MCMC Method |
The monotone data MCMC method was first proposed by Li (1988), and Liu (1993) described the algorithm. The method is useful especially when a data set is close to having a monotone missing pattern. In this case, the method needs to impute only a few missing values to the data set to have a monotone missing pattern in the imputed data set. Compared to a full data imputation that imputes all missing values, the monotone data MCMC method imputes fewer missing values in each iteration and achieves approximate stationarity in fewer iterations (Schafer 1997, p. 227).
You can request the monotone MCMC method by specifying the option IMPUTE=MONOTONE in the MCMC statement. The "Missing Data Patterns" table now denotes the variables with missing values by "." or "O". The value "." means that the variable is missing and will be imputed, and the value "O" means that the variable is missing and will not be imputed. The "Variance Information" and "Parameter Estimates" tables are not created.
You must specify the variables in the VAR statement. The variable order in the list determines the monotone missing pattern in the imputed data set. With a different order in the VAR list, the results will be different because the monotone missing pattern to be constructed will be different.
Assuming that the data are from a multivariate normal distribution, then like the MCMC method, the monotone MCMC method repeats the following steps:
1. The imputation I-step Given an estimated mean vector and covariance matrix, the I-step simulates the missing values for each observation independently. Only a subset of missing values are simulated to achieve a monotone pattern of missingness.
2. The posterior P-step Given a new sample with a monotone pattern of missingness, the P-step simulates the posterior population mean vector and covariance matrix with a noninformative Jeffreys prior. These new estimates are then used in the next I-step.
Imputation Step
The I-step is almost identical to the I-step described in the section MCMC Method for Arbitrary Missing Data except that only a subset of missing values need to be simulated. To state this precisely, denote the variables with observed values for observation i by 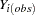 and the variables with missing values by
and the variables with missing values by  , where
, where  is a subset of the missing variables that will cause a monotone missingness when their values are imputed. Then the I-step draws values for
is a subset of the missing variables that will cause a monotone missingness when their values are imputed. Then the I-step draws values for  from a conditional distribution for
from a conditional distribution for  given
given 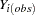 .
.
Posterior Step
The P-step is different from the P-step described in the section MCMC Method for Arbitrary Missing Data. Instead of simulating the 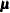 and
and 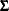 parameters from the full imputed data set, this P-step simulates the
parameters from the full imputed data set, this P-step simulates the 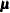 and
and 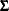 parameters through simulated regression coefficients from regression models based on the imputed data set with a monotone pattern of missingness. The step is similar to the process described in the section Regression Method for Monotone Missing Data.
parameters through simulated regression coefficients from regression models based on the imputed data set with a monotone pattern of missingness. The step is similar to the process described in the section Regression Method for Monotone Missing Data.
That is, for the variable 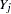 , a model
, a model
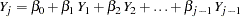 |
is fitted using  nonmissing observations for variable
nonmissing observations for variable 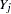 in the imputed data sets.
in the imputed data sets.
The fitted model consists of the regression parameter estimates 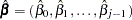 and the associated covariance matrix
and the associated covariance matrix  , where
, where 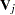 is the usual
is the usual 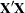 inverse matrix from the intercept and variables
inverse matrix from the intercept and variables 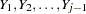 .
.
For each imputation, new parameters 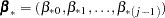 and
and 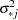 are drawn from the posterior predictive distribution of the parameters. That is, they are simulated from
are drawn from the posterior predictive distribution of the parameters. That is, they are simulated from 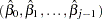 ,
, 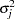 , and
, and 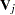 . The variance is drawn as
. The variance is drawn as
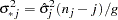 |
where  is a
is a 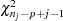 random variate and
random variate and  is the number of nonmissing observations for
is the number of nonmissing observations for 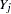 . The regression coefficients are drawn as
. The regression coefficients are drawn as
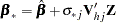 |
where 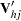 is the upper triangular matrix in the Cholesky decomposition,
is the upper triangular matrix in the Cholesky decomposition, 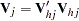 , and
, and 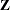 is a vector of
is a vector of 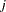 independent random normal variates.
independent random normal variates.
These simulated values of 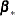 and
and 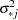 are then used to re-create the parameters
are then used to re-create the parameters 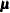 and
and 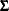 . For a detailed description of how to produce monotone missingness with the MCMC method for a multivariate normal data, see Schafer (1997, pp. 226–235).
. For a detailed description of how to produce monotone missingness with the MCMC method for a multivariate normal data, see Schafer (1997, pp. 226–235).
Copyright © 2009 by SAS Institute Inc., Cary, NC, USA. All rights reserved.