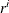The HPGENSELECT Procedure
-
Overview

- Getting Started
-
Syntax

-
Details
 Missing ValuesExponential Family DistributionsResponse DistributionsResponse Probability Distribution FunctionsLog-Likelihood FunctionsThe LASSO Method of Model SelectionUsing Validation and Test DataComputational Method: MultithreadingChoosing an Optimization AlgorithmDisplayed OutputODS Table Names
Missing ValuesExponential Family DistributionsResponse DistributionsResponse Probability Distribution FunctionsLog-Likelihood FunctionsThe LASSO Method of Model SelectionUsing Validation and Test DataComputational Method: MultithreadingChoosing an Optimization AlgorithmDisplayed OutputODS Table Names -
Examples

- References
PROC HPGENSELECT Statement
-
PROC HPGENSELECT <options>;
The PROC HPGENSELECT statement invokes the procedure. Table 8.1 summarizes the available options in the PROC HPGENSELECT statement by function. The options are then described fully in alphabetical order.
Table 8.1: PROC HPGENSELECT Statement Options
|
Option |
Description |
|---|---|
|
Basic Options |
|
|
Specifies a global significance level |
|
|
Specifies the input data set |
|
|
Limits the length of effect names |
|
|
Output Options |
|
|
Displays the "Parameter Estimates Correlation Matrix" table |
|
|
Displays the "Parameter Estimates Covariance Matrix" table |
|
|
Displays the "Iteration History" table when no model selection is performed |
|
|
Displays the summarized "Iteration History" table when model selection is performed |
|
|
Displays the summarized "Iteration History" table when no model selection is performed |
|
|
Suppresses ODS output |
|
|
Limits or suppresses the display of classification variable levels |
|
|
Suppresses computation of the covariance matrix and standard errors |
|
|
Optimization Options |
|
|
Tunes the absolute function convergence criterion |
|
|
Tunes the absolute function difference convergence criterion |
|
|
Tunes the absolute gradient convergence criterion |
|
|
Tunes the relative function difference convergence criterion |
|
|
Tunes the relative gradient convergence criterion |
|
|
Specifies the SAS data set that contains starting values, bounds, and constraints for single parameters |
|
|
Specifies the base regularization parameter for the LASSO method |
|
|
Specifies the maximum number of steps for the LASSO method |
|
|
Chooses the maximum number of iterations in any optimization |
|
|
Specifies the maximum number of function evaluations in any optimization |
|
|
Specifies the upper limit of CPU time (in seconds) for any optimization |
|
|
Specifies the minimum number of iterations in any optimization |
|
|
Specifies whether the objective function is normalized during optimization |
|
|
Adds parameter names to the "Parameter Estimates" table |
|
|
Selects the optimization technique |
|
|
Tolerance Options |
|
|
Specifies the convergence criterion for the LASSO method |
|
|
Tunes the singularity criterion for Cholesky decompositions |
|
|
Tunes the singularity criterion for the sweep operator |
|
|
Tunes the general singularity criterion |
|
|
User-Defined Format Options |
|
|
Specifies the file reference for a format stream |
|
You can specify the following options in the PROC HPGENSELECT statement.
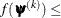
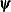
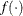
![\[ |f(\bpsi ^{(k-1)}) - f(\bpsi ^{(k)})| \leq r \]](images/stathpug_hpgenselect0006.png)
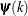
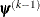
![\[ \max _ j |g_ j(\bpsi ^{(k)})| \leq r \]](images/stathpug_hpgenselect0009.png)
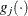
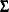
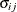
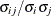
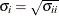
![\[ \frac{|f(\bpsi ^{(k)}) - f(\bpsi ^{(k-1)})|}{|f(\bpsi ^{(k-1)})|} \leq r \]](images/stathpug_hpgenselect0015.png)
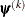
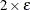

![\[ \frac{\mb{g}(\bpsi ^{(k)})^\prime [\bH ^{(k)}]^{-1} \mb{g}(\bpsi ^{(k)})}{|f(\bpsi ^{(k)})| } \leq r \]](images/stathpug_hpgenselect0019.png)
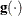
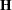
![\[ \frac{\parallel \mb{g}(\bpsi ^{(k)}) \parallel _2^2 \quad \parallel \mb{s}(\bpsi ^{(k)}) \parallel _2}{\parallel \mb{g}(\bpsi ^{(k)}) - \mb{g}(\bpsi ^{(k-1)}) \parallel _2 |f(\bpsi ^{(k)})| } \leq r \]](images/stathpug_hpgenselect0022.png)