The Decomposition Algorithm
- Overview
-
Getting Started

-
Syntax
 Decomposition Algorithm Options in the PROC OPTLP Statement or the SOLVE WITH LP Statement in PROC OPTMODELDecomposition Algorithm Options in the PROC OPTMILP Statement or the SOLVE WITH MILP Statement in PROC OPTMODELDECOMP StatementDECOMP_MASTER StatementDECOMP_MASTER_IP StatementDECOMP_SUBPROB Statement
Decomposition Algorithm Options in the PROC OPTLP Statement or the SOLVE WITH LP Statement in PROC OPTMODELDecomposition Algorithm Options in the PROC OPTMILP Statement or the SOLVE WITH MILP Statement in PROC OPTMODELDECOMP StatementDECOMP_MASTER StatementDECOMP_MASTER_IP StatementDECOMP_SUBPROB Statement -
Details

-
Examples
 Multicommodity Flow ProblemGeneralized Assignment ProblemBlock-Diagonal Structure and METHOD=AUTO in Single-Machine ModeBlock-Diagonal Structure and METHOD=AUTO in Distributed ModeBin Packing ProblemResource Allocation ProblemVehicle Routing ProblemATM Cash Management in Single-Machine ModeATM Cash Management in Distributed ModeKidney Donor Exchange
Multicommodity Flow ProblemGeneralized Assignment ProblemBlock-Diagonal Structure and METHOD=AUTO in Single-Machine ModeBlock-Diagonal Structure and METHOD=AUTO in Distributed ModeBin Packing ProblemResource Allocation ProblemVehicle Routing ProblemATM Cash Management in Single-Machine ModeATM Cash Management in Distributed ModeKidney Donor Exchange - References
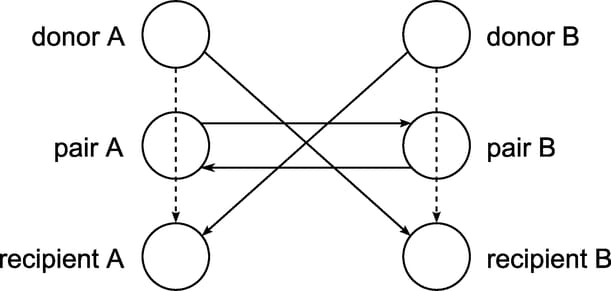
This example looks at an application of integer programming to help create a kidney donor exchange. Suppose someone needs a kidney transplant and a family member is willing to donate one. If the donor and recipient are incompatible (because of blood types, tissue mismatch, and so on), the transplant cannot happen. Now suppose two donor-recipient pairs A and B are in this situation, but donor A is compatible with recipient B and donor B is compatible with recipient A. Then two transplants can take place in a two-way swap, shown graphically in Output 14.10.1.
More generally, an ![]() -way swap that involves
-way swap that involves ![]() donors and
donors and ![]() recipients can be performed (Willingham 2009). To model this problem, define a directed graph as follows. Each node is an incompatible donor-recipient pair. Link
recipients can be performed (Willingham 2009). To model this problem, define a directed graph as follows. Each node is an incompatible donor-recipient pair. Link ![]() exists if the donor from node
exists if the donor from node ![]() is compatible with the recipient from node
is compatible with the recipient from node ![]() . Let
. Let ![]() define the set of nodes and
define the set of nodes and ![]() define the set of arcs. The link weight,
define the set of arcs. The link weight, ![]() , is a measure of the quality of the match. By introducing dummy links whose weight is 0, you can also include altruistic
donors who have no recipients, or recipients who have no donors. The idea is to find a maximum-weight node-disjoint union
of directed cycles. You want the union to be node-disjoint so that no kidney is donated more than once, and you want cycles
so that the donor from node
, is a measure of the quality of the match. By introducing dummy links whose weight is 0, you can also include altruistic
donors who have no recipients, or recipients who have no donors. The idea is to find a maximum-weight node-disjoint union
of directed cycles. You want the union to be node-disjoint so that no kidney is donated more than once, and you want cycles
so that the donor from node ![]() gives up a kidney if and only if the recipient from node
gives up a kidney if and only if the recipient from node ![]() receives a kidney.
receives a kidney.
Without any other constraints, the problem could be solved as a linear assignment problem. But doing so would allow arbitrarily
long cycles in the solution. Because of practical considerations (such as travel) and to mitigate risk, each cycle must have
no more than ![]() links. The kidney exchange problem is to find a maximum-weight node-disjoint union of short directed cycles.
links. The kidney exchange problem is to find a maximum-weight node-disjoint union of short directed cycles.
Define an index set ![]() of candidate disjoint unions of short cycles (called matchings). Let
of candidate disjoint unions of short cycles (called matchings). Let ![]() be a binary variable, which, if set to
be a binary variable, which, if set to ![]() , indicates that arc
, indicates that arc ![]() is in a matching
is in a matching ![]() . Let
. Let ![]() be a binary variable, which, if set to
be a binary variable, which, if set to ![]() , indicates that node
, indicates that node ![]() is covered by matching
is covered by matching ![]() . In addition, let
. In addition, let ![]() be a binary slack variable, which, if set to
be a binary slack variable, which, if set to ![]() , indicates that node
, indicates that node ![]() is not covered by any matching.
is not covered by any matching.
The kidney donor exchange can be formulated as a MILP as follows:
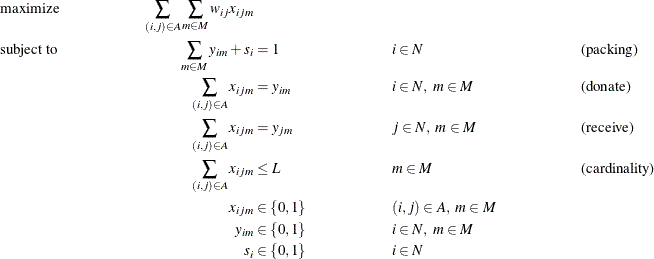
In this formulation, constraints (packing) ensure that each node is covered by at most one matching. Constraints (donate)
and (receive) enforce the condition that if node ![]() is covered by matching
is covered by matching ![]() , then the matching
, then the matching ![]() must use exactly one arc that leaves node
must use exactly one arc that leaves node ![]() (donate) and one arc that enters node
(donate) and one arc that enters node ![]() (receive). Conversely, if node
(receive). Conversely, if node ![]() is not covered by matching
is not covered by matching ![]() , then no arcs that enter or leave node
, then no arcs that enter or leave node ![]() can be used by matching
can be used by matching ![]() . Constraints (cardinality) enforce the condition that the number of arcs in matching
. Constraints (cardinality) enforce the condition that the number of arcs in matching ![]() must not exceed
must not exceed ![]() .
.
In this formulation, the matching identifier is arbitrary. Because it is not necessary to cover each incompatible donor-recipient
pair (node), the packing constraints can be modeled by using set partitioning constraints and the slack variable ![]() . Consider a decomposition by matching, where the packing constraints form the master problem and all other constraints form
identical matching subproblems. As described in the section Special Case: Identical Blocks, this is a situation in which an aggregate formulation and Ryan-Foster branching can greatly improve performance by reducing
symmetry.
. Consider a decomposition by matching, where the packing constraints form the master problem and all other constraints form
identical matching subproblems. As described in the section Special Case: Identical Blocks, this is a situation in which an aggregate formulation and Ryan-Foster branching can greatly improve performance by reducing
symmetry.
The following DATA step sets up the problem, first creating a random graph on ![]() nodes with link probability
nodes with link probability ![]() and Uniform(0,1) weight:
and Uniform(0,1) weight:
/* create random graph on n nodes with arc probability p
and uniform(0,1) weight */
%let n = 100;
%let p = 0.02;
data ArcData;
do i = 0 to &n - 1;
do j = 0 to &n - 1;
if i eq j then continue;
else if ranuni(1) < &p then do;
weight = ranuni(2);
output;
end;
end;
end;
run;
The following PROC OPTMODEL statements read in the data, declare the optimization model, and use the decomposition algorithm to solve it:
%let max_length = 10;
proc optmodel;
set <num,num> ARCS;
num weight {ARCS};
read data ArcData into ARCS=[i j] weight;
print weight;
set NODES = union {<i,j> in ARCS} {i,j};
set MATCHINGS = 1..card(NODES)/2;
/* UseNode[i,m] = 1 if node i is used in matching m, 0 otherwise */
var UseNode {NODES, MATCHINGS} binary;
/* UseArc[i,j,m] = 1 if arc (i,j) is used in matching m, 0 otherwise */
var UseArc {ARCS, MATCHINGS} binary;
/* maximize total weight of arcs used */
max TotalWeight
= sum {<i,j> in ARCS, m in MATCHINGS} weight[i,j] * UseArc[i,j,m];
/* each node appears in at most one matching */
/* rewrite as set partitioning (so decomp uses identical blocks)
sum{} x <= 1 => sum{} x + s = 1, s >= 0 with no associated cost */
var Slack {NODES} binary;
con node_packing {i in NODES}:
sum {m in MATCHINGS} UseNode[i,m] + Slack[i] = 1;
/* at most one donee for each donor */
con donate {i in NODES, m in MATCHINGS}:
sum {<(i),j> in ARCS} UseArc[i,j,m] = UseNode[i,m];
/* at most one donor for each donee */
con receive {j in NODES, m in MATCHINGS}:
sum {<i,(j)> in ARCS} UseArc[i,j,m] = UseNode[j,m];
/* exclude long matchings */
con cardinality {m in MATCHINGS}:
sum {<i,j> in ARCS} UseArc[i,j,m] <= &max_length;
/* decompose by matching (aggregate formulation) */
for {i in NODES, m in MATCHINGS} donate[i,m].block = m;
for {j in NODES, m in MATCHINGS} receive[j,m].block = m;
for {m in MATCHINGS} cardinality[m].block = m;
solve with milp / presolver=basic decomp=();
/* save solution to a data set */
create data Solution from
[m i j]={m in MATCHINGS, <i,j> in ARCS: UseArc[i,j,m].sol > 0.5}
weight[i,j];
quit;
In this case, the PRESOLVER=BASIC option ensures that the model maintains its specified symmetry, enabling the algorithm to use the aggregate formulation and Ryan-Foster branching. The solution summary is displayed in Output 14.10.2.
Output 14.10.2: Solution Summary
| Solution Summary | |
|---|---|
| Solver | MILP |
| Algorithm | Decomposition |
| Objective Function | TotalWeight |
| Solution Status | Optimal |
| Objective Value | 26.020287142 |
| Relative Gap | 0 |
| Absolute Gap | 0 |
| Primal Infeasibility | 2.997602E-15 |
| Bound Infeasibility | 1.110223E-15 |
| Integer Infeasibility | 2.775558E-15 |
| Best Bound | 26.020287142 |
| Nodes | 25 |
| Iterations | 155 |
| Presolve Time | 0.17 |
| Solution Time | 24.51 |
The iteration log is displayed in Output 14.10.3.
Output 14.10.3: Log
| NOTE: There were 194 observations read from the data set WORK.ARCDATA. |
| NOTE: Problem generation will use 4 threads. |
| NOTE: The problem has 14065 variables (0 free, 0 fixed). |
| NOTE: The problem has 14065 binary and 0 integer variables. |
| NOTE: The problem has 9457 linear constraints (48 LE, 9409 EQ, 0 GE, 0 range). |
| NOTE: The problem has 42001 linear constraint coefficients. |
| NOTE: The problem has 0 nonlinear constraints (0 LE, 0 EQ, 0 GE, 0 range). |
| NOTE: The MILP presolver value BASIC is applied. |
| NOTE: The MILP presolver removed 4786 variables and 3298 constraints. |
| NOTE: The MILP presolver removed 14290 constraint coefficients. |
| NOTE: The MILP presolver modified 0 constraint coefficients. |
| NOTE: The presolved problem has 9279 variables, 6159 constraints, and 27711 |
| constraint coefficients. |
| NOTE: The MILP solver is called. |
| NOTE: The Decomposition algorithm is used. |
| NOTE: The Decomposition algorithm is executing in single-machine mode. |
| NOTE: The DECOMP method value USER is applied. |
| NOTE: All blocks are identical. |
| NOTE: The Decomposition algorithm is using an aggregate formulation and Ryan-Foster |
| branching. |
| NOTE: The problem has a decomposable structure with 48 blocks. The largest block |
| covers 2.06% of the constraints in the problem. |
| NOTE: The decomposition subproblems cover 9216 (99.32%) variables and 6096 (98.98%) |
| constraints. |
| NOTE: The deterministic parallel mode is enabled. |
| NOTE: The Decomposition algorithm is using up to 4 threads. |
| Iter Best Master Best LP IP CPU Real |
| Bound Objective Integer Gap Gap Time Time |
| NOTE: Starting phase 1. |
| 1 0.0000 0.0000 . 0.00% . 0 0 |
| NOTE: Starting phase 2. |
| . 390.3703 10.9852 10.9852 97.19% 97.19% 0 0 |
| 2 390.2342 10.9852 10.9852 97.18% 97.18% 0 0 |
| 3 380.7461 10.9852 10.9852 97.11% 97.11% 0 0 |
| 4 353.1404 10.9852 10.9852 96.89% 96.89% 0 0 |
| 5 344.6969 16.9959 16.9959 95.07% 95.07% 0 0 |
| 7 337.0188 17.7608 16.9959 94.73% 94.96% 0 0 |
| 9 317.4067 19.1811 16.9959 93.96% 94.65% 0 0 |
| . 317.4067 21.1135 21.1135 93.35% 93.35% 0 0 |
| 10 317.4067 21.1135 21.1135 93.35% 93.35% 0 0 |
| 11 258.8116 21.4365 21.1135 91.72% 91.84% 0 0 |
| 13 188.1070 23.4494 21.1135 87.53% 88.78% 0 0 |
| 16 154.3908 24.6477 21.1135 84.04% 86.32% 0 0 |
| 17 132.1716 24.7011 21.1135 81.31% 84.03% 0 0 |
| 18 126.9697 24.8435 21.1135 80.43% 83.37% 0 1 |
| 19 84.6317 24.9162 21.1135 70.56% 75.05% 1 1 |
| . 84.6317 24.9510 21.1778 70.52% 74.98% 1 1 |
| 20 84.6317 24.9510 21.1778 70.52% 74.98% 1 1 |
| 28 71.2689 26.4398 21.1778 62.90% 70.28% 1 1 |
| . 71.2689 26.6775 22.2734 62.57% 68.75% 2 2 |
| 30 46.3628 26.6775 22.2734 42.46% 51.96% 2 2 |
| 32 43.4466 26.7648 22.2734 38.40% 48.73% 2 2 |
| 33 34.2503 26.7648 22.2734 21.86% 34.97% 2 2 |
| 36 33.1439 26.7804 22.2734 19.20% 32.80% 3 3 |
| 37 29.0201 26.7804 22.2734 7.72% 23.25% 3 3 |
| 38 26.7804 26.7804 22.2734 0.00% 16.83% 3 3 |
| . 26.7804 26.7804 22.3657 0.00% 16.48% 3 3 |
| 38 26.7804 26.7804 22.3657 0.00% 16.48% 3 3 |
| NOTE: Starting branch and bound. |
| Node Active Sols Best Best Gap CPU Real |
| Integer Bound Time Time |
| 0 1 9 22.3657 26.7804 16.48% 3 3 |
| 5 7 10 24.6832 26.4468 6.67% 11 11 |
| 10 12 11 24.6832 26.3728 6.41% 13 13 |
| 14 8 12 25.9769 26.2093 0.89% 16 16 |
| 15 9 13 25.9955 26.2093 0.82% 16 17 |
| 19 5 15 26.0203 26.1602 0.53% 19 20 |
| 20 4 15 26.0203 26.0993 0.30% 19 20 |
| 24 0 15 26.0203 26.0203 0.00% 22 23 |
| NOTE: The Decomposition algorithm used 1 threads. |
| NOTE: The Decomposition algorithm time is 23.04 seconds. |
| NOTE: Optimal. |
| NOTE: Objective = 26.020287142. |
| NOTE: The data set WORK.SOLUTION has 42 observations and 4 variables. |
The solution is a set of arcs that define a union of short directed cycles (matchings). The following call to PROC OPTNET
extracts the corresponding cycles from the list of arcs and outputs them to the data set Cycles:
proc optnet
direction = directed
data_links = Solution;
data_links_var
from = i
to = j;
cycle
mode = all_cycles
out = Cycles;
run;
For more information about PROC OPTNET, see
SAS/OR User's Guide: Network Optimization Algorithms. Alternatively, you can extract the cycles by using the SOLVE WITH NETWORK statement in PROC OPTMODEL (see Chapter 8: The Network Solver). The optimal donor exchanges from the output data set Cycles are displayed in Output 14.10.4.
Output 14.10.4: Optimal Donor Exchanges
| order | node |
|---|---|
| 1 | 5 |
| 2 | 19 |
| 3 | 56 |
| 4 | 12 |
| 5 | 33 |
| 6 | 70 |
| 7 | 63 |
| 8 | 43 |
| 9 | 15 |
| 10 | 5 |
| order | node |
|---|---|
| 1 | 13 |
| 2 | 74 |
| 3 | 65 |
| 4 | 41 |
| 5 | 59 |
| 6 | 50 |
| 7 | 49 |
| 8 | 98 |
| 9 | 13 |
| order | node |
|---|---|
| 1 | 16 |
| 2 | 91 |
| 3 | 17 |
| 4 | 57 |
| 5 | 87 |
| 6 | 72 |
| 7 | 64 |
| 8 | 22 |
| 9 | 88 |
| 10 | 16 |
| order | node |
|---|---|
| 1 | 8 |
| 2 | 32 |
| 3 | 79 |
| 4 | 71 |
| 5 | 69 |
| 6 | 26 |
| 7 | 9 |
| 8 | 18 |
| 9 | 95 |
| 10 | 35 |
| 11 | 8 |
| order | node |
|---|---|
| 1 | 52 |
| 2 | 77 |
| 3 | 94 |
| 4 | 81 |
| 5 | 52 |
| order | node |
|---|---|
| 1 | 24 |
| 2 | 92 |
| 3 | 24 |