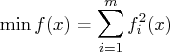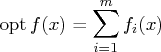| The NLP Procedure |
Covariance Matrix
The COV= option must be specified to compute an approximate covariance matrix for the parameter estimates under asymptotic theory for least-squares, maximum-likelihood, or Bayesian estimation, with or without corrections for degrees of freedom as specified by the VARDEF= option.
Two groups of six different forms of covariance matrices (and therefore approximate standard errors) can be computed corresponding to the following two situations:
- The LSQ statement is specified, which means that
least-squares estimates are being computed:
- The MIN or MAX statement is specified, which means
that maximum-likelihood or Bayesian estimates are
being computed:
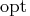 is either
is either 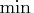 or
or 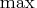 .
.
In either case, the following matrices are used:
For unconstrained minimization, or when none of the final parameter estimates are subjected to linear equality or active inequality constraints, the formulas of the six types of covariance matrices are as follows:
| COV | MIN or MAX Statement | LSQ Statement | |
| 1 | M | ||
| 2 | H | ||
| 3 | J | ||
| 4 | B | ||
| 5 | E | ||
| 6 | U | ||
The value of ![]() depends on the VARDEF= option and on the value
of the _NOBS_ variable:
depends on the VARDEF= option and on the value
of the _NOBS_ variable:
The value ![]() depends on the specification of the SIGSQ=
option and on the value of
depends on the specification of the SIGSQ=
option and on the value of ![]() :
:
The two groups of formulas distinguish between two situations:
- For least-squares estimates, the error variance can be estimated from the objective function value and is used in three of the six different forms of covariance matrices. If you have an independent estimate of the error variance, you can specify it with the SIGSQ= option.
- For maximum-likelihood or Bayesian estimates, the objective function should be the logarithm of the likelihood or of the posterior density when using the MAX statement.
For minimization, the inversion of the matrices in these formulas is done so that negative eigenvalues are considered zero, resulting always in a positive semidefinite covariance matrix.
In small samples, estimates of the covariance matrix based on asymptotic theory are often too small and should be used with caution.
If the final parameter estimates are subjected to ![]() linear equality or active linear inequality constraints, the
formulas of the covariance matrices are modified
similar to Gallant (1987) and Cramer (1986, p. 38) and
additionally generalized for applications with singular
matrices. In the constrained case, the value of
linear equality or active linear inequality constraints, the
formulas of the covariance matrices are modified
similar to Gallant (1987) and Cramer (1986, p. 38) and
additionally generalized for applications with singular
matrices. In the constrained case, the value of ![]() used in
the scalar factor
used in
the scalar factor ![]() is defined by
is defined by
For minimization, the covariance matrix should be positive definite; for maximization it should be negative definite. There are several options available to check for a rank deficiency of the covariance matrix:
- The ASINGULAR=, MSINGULAR=,
and VSINGULAR= options can
be used to set three singularity criteria for the inversion
of the matrix
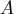 needed to compute the covariance matrix,
when
needed to compute the covariance matrix,
when 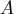 is either the Hessian
or one of the crossproduct Jacobian matrices.
The singularity criterion used for the inversion is
is either the Hessian
or one of the crossproduct Jacobian matrices.
The singularity criterion used for the inversion is
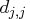 is the diagonal pivot of the matrix
is the diagonal pivot of the matrix 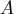 ,
and ASING, VSING and MSING are the specified values of
the ASINGULAR=, VSINGULAR=,
and MSINGULAR= options. The default values are
,
and ASING, VSING and MSING are the specified values of
the ASINGULAR=, VSINGULAR=,
and MSINGULAR= options. The default values are
- ASING: the square root of the smallest positive double precision value
- MSING: 1E-12 if the SINGULAR= option is not specified
and
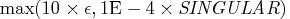 otherwise,
where
otherwise,
where 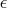 is the machine precision
is the machine precision
- VSING: 1E-8 if the SINGULAR= option is not specified and the value of SINGULAR otherwise
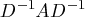 is decomposed and the singularity criteria are modified
correspondingly.
is decomposed and the singularity criteria are modified
correspondingly.
- If the matrix
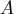 is found singular in the first step,
a generalized inverse is computed. Depending on the G4=
option, a generalized inverse is computed that satisfies either
all four or only two Moore-Penrose conditions.
If the number of parameters
is found singular in the first step,
a generalized inverse is computed. Depending on the G4=
option, a generalized inverse is computed that satisfies either
all four or only two Moore-Penrose conditions.
If the number of parameters 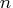 of the
application is less than or equal to G4=
of the
application is less than or equal to G4=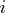 , a G4 inverse
is computed; otherwise only a G2 inverse is computed.
The G4 inverse is computed by (the computationally very
expensive but numerically stable) eigenvalue decomposition;
the G2 inverse is computed by Gauss transformation.
The G4 inverse is computed using the eigenvalue
decomposition
, a G4 inverse
is computed; otherwise only a G2 inverse is computed.
The G4 inverse is computed by (the computationally very
expensive but numerically stable) eigenvalue decomposition;
the G2 inverse is computed by Gauss transformation.
The G4 inverse is computed using the eigenvalue
decomposition 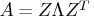 , where
, where 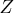 is the
orthogonal matrix of eigenvectors and
is the
orthogonal matrix of eigenvectors and 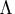 is
the diagonal matrix of eigenvalues,
is
the diagonal matrix of eigenvalues,
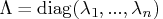 .
If the PEIGVAL option is specified, the eigenvalues
.
If the PEIGVAL option is specified, the eigenvalues
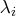 are displayed. The G4 inverse of
are displayed. The G4 inverse of 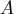 is set to
is set to
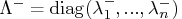 is defined using the COVSING= option:
is defined using the COVSING= option:
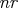 smallest eigenvalues are set to zero, where
smallest eigenvalues are set to zero, where 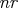 is
the number of rank deficiencies found in the first step.
is
the number of rank deficiencies found in the first step.
For optimization techniques that do not use second-order derivatives, the covariance matrix is usually computed using finite-difference approximations of the derivatives. By specifying TECH=NONE, any of the covariance matrices can be computed using analytical derivatives. The covariance matrix specified by the COV= option can be displayed (using the PCOV option) and is written to the OUTEST= data set.
Copyright © 2008 by SAS Institute Inc., Cary, NC, USA. All rights reserved.