The following example estimates the normal SSM of the mink-muskrat data by using the EM algorithm. The mink-muskrat series are detrended. Refer to Harvey (1989) for details of this data set. Since this EM algorithm uses filtering and smoothing, you can learn how to use the KALCVF and KALCVS calls to analyze the data. Consider the bivariate SSM:
where ![]() is a
is a ![]() identity matrix, the observation noise has a time-invariant covariance matrix
identity matrix, the observation noise has a time-invariant covariance matrix ![]() , and the covariance matrix of the transition equation is also assumed to be time invariant. The initial state
, and the covariance matrix of the transition equation is also assumed to be time invariant. The initial state ![]() has mean
has mean ![]() and covariance
and covariance ![]() . For estimation, the
. For estimation, the ![]() matrix is fixed as
matrix is fixed as
while the mean vector ![]() is updated by the smoothing procedure such that
is updated by the smoothing procedure such that ![]() . Note that this estimation requires an extra smoothing step since the usual smoothing procedure does not produce
. Note that this estimation requires an extra smoothing step since the usual smoothing procedure does not produce ![]() .
.
The EM algorithm maximizes the expected log-likelihood function given the current parameter estimates. In practice, the log-likelihood function of the normal SSM is evaluated while the parameters are updated by using the M-step of the EM maximization
![\begin{eqnarray*} \bF ^{i+1} & = & \bS _ t(1)[\bS _{t-1}(0)]^{-1} \\ \bV ^{i+1} & = & \frac{1}{T} \left( \bS _ t(0) - \bS _ t(1)[\bS _{t-1}(0)]^{-1} \bS ^{\prime }_ t(1) \right) \\ \bR ^{i+1} & = & \frac{1}{T} \sum _{t=1}^ T \left[ (\textbf{y}_ t - \bH \textbf{z}_{t|T}) (\textbf{y}_ t - \bH \textbf{z}_{t|T})^{\prime } + \bH \bP _{t|T} \bH ^{\prime } \right] \\ \mu ^{i+1} & = & \textbf{z}_{0|T} \end{eqnarray*}](images/imlug_timeseriesexpls0446.png)
where the index ![]() represents the current iteration number, and
represents the current iteration number, and
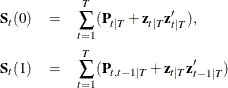
It is necessary to compute the value of ![]() recursively such that
recursively such that
where ![]() and the initial value
and the initial value ![]() is derived by using the formula
is derived by using the formula
Note that the initial value of the state vector is updated for each iteration
The objective function value is computed as ![]() in the IML module LIK. The log-likelihood function is written
in the IML module LIK. The log-likelihood function is written
where ![]() .
.
The iteration history is shown in Output 13.3.1. As shown in Output 13.3.2, the eigenvalues of ![]() are within the unit circle, which indicates that the SSM is stationary. However, the muskrat series (Y1) is reported to be
difference stationary. The estimated parameters are almost identical to those of the VAR(1) estimates. Refer to Harvey (1989). Finally, multistep forecasts of
are within the unit circle, which indicates that the SSM is stationary. However, the muskrat series (Y1) is reported to be
difference stationary. The estimated parameters are almost identical to those of the VAR(1) estimates. Refer to Harvey (1989). Finally, multistep forecasts of ![]() are computed by using the KALCVF call. Here is the code:
are computed by using the KALCVF call. Here is the code:
call kalcvf(pred,vpred,filt,vfilt,y,15,a,f,b,h,var,z0,vz0);
The predicted values of the state vector ![]() and their standard errors are shown in Output 13.3.3. Here is the code:
and their standard errors are shown in Output 13.3.3. Here is the code:
title 'SSM Estimation using EM Algorithm';
data one;
input y1 y2 @@;
datalines;
0.10609 0.16794 -0.16852 0.06242 -0.23700 -0.13344
-0.18022 -0.50616 0.18094 -0.37943 0.65983 -0.40132
0.65235 0.08789 0.21594 0.23877 -0.11515 0.40043
-0.00067 0.37758 -0.00387 0.55735 -0.25202 0.34444
-0.65011 -0.02749 -0.53646 -0.41519 -0.08462 0.02591
-0.05640 -0.11348 0.26630 0.20544 0.03641 0.16331
-0.26030 -0.01498 -0.03995 0.09657 0.33612 0.31096
-0.11672 0.30681 -0.69775 -0.69351 -0.07569 -0.56212
0.36149 -0.36799 0.42341 -0.24725 0.26721 0.04478
-0.00363 0.21637 0.08333 0.30188 -0.22480 0.29493
-0.13728 0.35463 -0.12698 0.05490 -0.18770 -0.52573
0.34741 -0.49541 0.54947 -0.26250 0.57423 -0.21936
0.57493 -0.12012 0.28188 0.63556 -0.58438 0.27067
-0.50236 0.10386 -0.60766 0.36748 -1.04784 -0.33493
-0.68857 -0.46525 -0.11450 -0.63648 0.22005 -0.26335
0.36533 0.07017 -0.00151 -0.04977 0.03740 -0.02411
0.22438 0.30790 -0.16196 0.41050 -0.12862 0.34929
0.08448 -0.14995 0.17945 -0.03320 0.37502 0.02953
0.95727 0.24090 0.86188 0.41096 0.39464 0.24157
0.53794 0.29385 0.13054 0.39336 -0.39138 -0.00323
-1.23825 -0.56953 -0.66286 -0.72363
;
run;
proc iml;
start lik(y,pred,vpred,h,rt);
n = nrow(y);
nz = ncol(h);
et = y - pred*h`;
sum1 = 0;
sum2 = 0;
do i = 1 to n;
vpred_i = vpred[(i-1)*nz+1:i*nz,];
et_i = et[i,];
ft = h*vpred_i*h` + rt;
sum1 = sum1 + log(det(ft));
sum2 = sum2 + et_i*inv(ft)*et_i`;
end;
return(sum1+sum2);
finish;
use one;
read all into y var {y1 y2};
close one;
/*-- mean adjust series --*/
t = nrow(y);
ny = ncol(y);
nz = ny;
f = i(nz);
h = i(ny);
/*-- observation noise variance is diagonal --*/
rt = 1e-5#i(ny);
/*-- transition noise variance --*/
vt = .1#i(nz);
a = j(nz,1,0);
b = j(ny,1,0);
myu = j(nz,1,0);
sigma = .1#i(nz);
converge = 0;
logl0 = 0.0;
do iter = 1 to 100 while( converge = 0 );
/*--- construct big cov matrix --*/
var = ( vt || j(nz,ny,0) ) //
( j(ny,nz,0) || rt );
/*-- initial values are changed --*/
z0 = myu` * f`;
vz0 = f * sigma * f` + vt;
/*-- filtering to get one-step prediction and filtered value --*/
call kalcvf(pred,vpred,filt,vfilt,y,0,a,f,b,h,var,z0,vz0);
/*-- smoothing using one-step prediction values --*/
call kalcvs(sm,vsm,y,a,f,b,h,var,pred,vpred);
/*-- compute likelihood values --*/
logl = lik(y,pred,vpred,h,rt);
/*-- store old parameters and function values --*/
myu0 = myu;
f0 = f;
vt0 = vt;
rt0 = rt;
diflog = logl - logl0;
logl0 = logl;
itermat = itermat // ( iter || logl0 || shape(f0,1) || myu0` );
/*-- obtain P*(t) to get P_T_0 and Z_T_0 --*/
/*-- these values are not usually needed --*/
/*-- See Harvey (1989 p154) or Shumway (1988, p177) --*/
jt1 = sigma * f` * inv(vpred[1:nz,]);
p_t_0 = sigma + jt1*(vsm[1:nz,] - vpred[1:nz,])*jt1`;
z_t_0 = myu + jt1*(sm[1,]` - pred[1,]`);
p_t1_t = vpred[(t-1)*nz+1:t*nz,];
p_t1_t1 = vfilt[(t-2)*nz+1:(t-1)*nz,];
kt = p_t1_t*h`*inv(h*p_t1_t*h`+rt);
/*-- obtain P_T_TT1. See Shumway (1988, p180) --*/
p_t_ii1 = (i(nz)-kt*h)*f*p_t1_t1;
st0 = vsm[(t-1)*nz+1:t*nz,] + sm[t,]`*sm[t,];
st1 = p_t_ii1 + sm[t,]`*sm[t-1,];
st00 = p_t_0 + z_t_0 * z_t_0`;
cov = (y[t,]` - h*sm[t,]`) * (y[t,]` - h*sm[t,]`)` +
h*vsm[(t-1)*nz+1:t*nz,]*h`;
do i = t to 2 by -1;
p_i1_i1 = vfilt[(i-2)*nz+1:(i-1)*nz,];
p_i1_i = vpred[(i-1)*nz+1:i*nz,];
jt1 = p_i1_i1 * f` * inv(p_i1_i);
p_i1_i = vpred[(i-2)*nz+1:(i-1)*nz,];
if ( i > 2 ) then
p_i2_i2 = vfilt[(i-3)*nz+1:(i-2)*nz,];
else
p_i2_i2 = sigma;
jt2 = p_i2_i2 * f` * inv(p_i1_i);
p_t_i1i2 = p_i1_i1*jt2` + jt1*(p_t_ii1 - f*p_i1_i1)*jt2`;
p_t_ii1 = p_t_i1i2;
temp = vsm[(i-2)*nz+1:(i-1)*nz,];
sm1 = sm[i-1,]`;
st0 = st0 + ( temp + sm1 * sm1` );
if ( i > 2 ) then
st1 = st1 + ( p_t_ii1 + sm1 * sm[i-2,]);
else st1 = st1 + ( p_t_ii1 + sm1 * z_t_0`);
st00 = st00 + ( temp + sm1 * sm1` );
cov = cov + ( h * temp * h` +
(y[i-1,]` - h * sm1)*(y[i-1,]` - h * sm1)` );
end;
/*-- M-step: update the parameters --*/
myu = z_t_0;
f = st1 * inv(st00);
vt = (st0 - st1 * inv(st00) * st1`)/t;
rt = cov / t;
/*-- check convergence --*/
if ( max(abs((myu - myu0)/(myu0+1e-6))) < 1e-2 &
max(abs((f - f0)/(f0+1e-6))) < 1e-2 &
max(abs((vt - vt0)/(vt0+1e-6))) < 1e-2 &
max(abs((rt - rt0)/(rt0+1e-6))) < 1e-2 &
abs((diflog)/(logl0+1e-6)) < 1e-3 ) then
converge = 1;
end;
reset noname;
colnm = {'Iter' '-2*log L' 'F11' 'F12' 'F21' 'F22'
'MYU11' 'MYU22'};
print itermat[colname=colnm format=8.4];
eval = eigval(f0);
colnm = {'Real' 'Imag' 'MOD'};
eval = eval || sqrt((eval#eval)[,+]);
print eval[colname=colnm];
var = ( vt || j(nz,ny,0) ) //
( j(ny,nz,0) || rt );
/*-- initial values are changed --*/
z0 = myu` * f`;
vz0 = f * sigma * f` + vt;
free itermat;
/*-- multistep prediction --*/
call kalcvf(pred,vpred,filt,vfilt,y,15,a,f,b,h,var,z0,vz0);
do i = 1 to 15;
itermat = itermat // ( i || pred[t+i,] ||
sqrt(vecdiag(vpred[(t+i-1)*nz+1:(t+i)*nz,]))` );
end;
colnm = {'n-Step' 'Z1_T_n' 'Z2_T_n' 'SE_Z1' 'SE_Z2'};
print itermat[colname=colnm];
quit;
Output 13.3.1: Iteration History
| SSM Estimation using EM Algorithm |
| Iter | -2*log L | F11 | F12 | F21 | F22 | MYU11 | MYU22 |
|---|---|---|---|---|---|---|---|
| 1.0000 | -154.010 | 1.0000 | 0.0000 | 0.0000 | 1.0000 | 0.0000 | 0.0000 |
| 2.0000 | -237.962 | 0.7952 | -0.6473 | 0.3263 | 0.5143 | 0.0530 | 0.0840 |
| 3.0000 | -238.083 | 0.7967 | -0.6514 | 0.3259 | 0.5142 | 0.1372 | 0.0977 |
| 4.0000 | -238.126 | 0.7966 | -0.6517 | 0.3259 | 0.5139 | 0.1853 | 0.1159 |
| 5.0000 | -238.143 | 0.7964 | -0.6519 | 0.3257 | 0.5138 | 0.2143 | 0.1304 |
| 6.0000 | -238.151 | 0.7963 | -0.6520 | 0.3255 | 0.5136 | 0.2324 | 0.1405 |
| 7.0000 | -238.153 | 0.7962 | -0.6520 | 0.3254 | 0.5135 | 0.2438 | 0.1473 |
| 8.0000 | -238.155 | 0.7962 | -0.6521 | 0.3253 | 0.5135 | 0.2511 | 0.1518 |
| 9.0000 | -238.155 | 0.7962 | -0.6521 | 0.3253 | 0.5134 | 0.2558 | 0.1546 |
| 10.0000 | -238.155 | 0.7961 | -0.6521 | 0.3253 | 0.5134 | 0.2588 | 0.1565 |
Output 13.3.2: Eigenvalues of F Matrix
| Real | Imag | MOD |
|---|---|---|
| 0.6547534 | 0.438317 | 0.7879237 |
| 0.6547534 | -0.438317 | 0.7879237 |
Output 13.3.3: Multistep Prediction
| n-Step | Z1_T_n | Z2_T_n | SE_Z1 | SE_Z2 |
|---|---|---|---|---|
| 1 | -0.055792 | -0.587049 | 0.2437666 | 0.237074 |
| 2 | 0.3384325 | -0.319505 | 0.3140478 | 0.290662 |
| 3 | 0.4778022 | -0.053949 | 0.3669731 | 0.3104052 |
| 4 | 0.4155731 | 0.1276996 | 0.4021048 | 0.3218256 |
| 5 | 0.2475671 | 0.2007098 | 0.419699 | 0.3319293 |
| 6 | 0.0661993 | 0.1835492 | 0.4268943 | 0.3396153 |
| 7 | -0.067001 | 0.1157541 | 0.430752 | 0.3438409 |
| 8 | -0.128831 | 0.0376316 | 0.4341532 | 0.3456312 |
| 9 | -0.127107 | -0.022581 | 0.4369411 | 0.3465325 |
| 10 | -0.086466 | -0.052931 | 0.4385978 | 0.3473038 |
| 11 | -0.034319 | -0.055293 | 0.4393282 | 0.3479612 |
| 12 | 0.0087379 | -0.039546 | 0.4396666 | 0.3483717 |
| 13 | 0.0327466 | -0.017459 | 0.439936 | 0.3485586 |
| 14 | 0.0374564 | 0.0016876 | 0.4401753 | 0.3486415 |
| 15 | 0.0287193 | 0.0130482 | 0.440335 | 0.3487034 |