The MODEL statement specifies the regression model and the error structure assumed for the regression residuals. The response
variable on the left side of the equal sign is regressed on the independent variables listed after the equal sign. Any number
of MODEL statements can be used. For each model statement, only one response variable can be specified on the left side of
the equal sign.
The error structure is specified by the FIXONE, FIXTWO, RANONE, RANTWO, FULLER, PARKS, and DASILVA options. More than one
of these options can be used, in which case the analysis is repeated for each error structure model specified.
Models can be given labels up to 32 characters in length. Model labels are used in the printed output to identify the results
for different models. If no label is specified, the response variable name is used as the label for the model. The model label
is specified as follows:
label:
The following options can be specified on the MODEL statement after a slash (/).
-
CORRB
-
-
CORR
-
prints the matrix of estimated correlations between the parameter estimates.
-
COVB
-
-
VAR
-
prints the matrix of estimated covariances between the parameter estimates.
-
FIXONE
-
specifies that a one-way fixed-effects model be estimated with the one-way model that corresponds to group effects only.
-
FIXTWO
-
specifies that a two-way fixed-effects model be estimated.
-
RANONE
-
specifies that a one-way random-effects model be estimated.
-
RANTWO
-
specifies that a two-way random-effects model be estimated.
-
FULLER
-
specifies that the model be estimated by using the Fuller-Battese method, which assumes a variance components model for the
error structure.
-
PARKS
-
specifies that the model be estimated by using the Parks method, which assumes a first-order autoregressive model for the
error structure.
-
DASILVA
-
specifies that the model be estimated by using the Da Silva method, which assumes a mixed variance-component moving-average
model for the error structure.
-
M=number
-
specifies the order of the moving-average process in the Da Silva method. The M= value must be less than  . The default is M=1.
. The default is M=1.
-
PHI
-
prints the 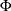 matrix of estimated covariances of the observations for the Parks method. The PHI option is relevant only when the PARKS
option is used.
matrix of estimated covariances of the observations for the Parks method. The PHI option is relevant only when the PARKS
option is used.
-
RHO
-
prints the estimated autocorrelation coefficients for the Parks method.
-
NOINT
-
-
NOMEAN
-
suppresses the intercept parameter from the model.
-
NOPRINT
-
suppresses the normal printed output.
-
SINGULAR=number
-
specifies a singularity criterion for the inversion of the matrix. The default depends on the precision of the computer system.