| What's New |
What's New in SAS/Genetics 9.2
| Overview |
SAS/Genetics includes two new experimental procedures, the BTL procedure and the GENESELECT procedure.
Several enhancements have been made to the ALLELE procedure, and a format of genotype columns not previously supported can now be accommodated by the ALLELE, CASECONTROL, FAMILY, and HAPLOTYPE procedures.
| Accommodating a New Data Format |
If your genotypes are represented by one character for each of their two alleles with no delimiting character separating them (such as "AB"), the GENOCOL and DELIMITER='' options allow the inclusion of such columns in the VAR statement of the ALLELE, CASECONTROL, FAMILY, and HAPLOTYPE procedures. Note that there is no space between the two quotation marks in the DELIMITER= option.
| ALLELE Procedure |
The new POP statement enables you to specify a variable that defines populations. Various 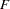 statistics can be computed that serve to describe the genetic structure of the population hierarchy.
statistics can be computed that serve to describe the genetic structure of the population hierarchy.
The MAXDIST= option of the PROC ALLELE statement can now be specified in terms of the unit used to define markers’ locations. A LOCATION variable can be included in the NDATA= data set, containing a numeric value that represents each marker’s location, and the MAXDIST= option is applied to the distance between markers (the absolute value of the difference of the two LOCATION values) for determining whether linkage disequilibrium (LD) measures are to be calculated for that particular pair. When the NDATA= option is not specified or the data set does not contain a LOCATION variable, the MAXDIST= option functions as it did previously, calculating the distance between markers as the number of markers apart they are.
The upper bound for the LD measure 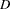 , used in the denominator of the
, used in the denominator of the 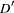 measure, is now calculated according to Hamilton and Cole (2004) and Zaykin (2004) when HAPLO=NONE or NONEHWD.
measure, is now calculated according to Hamilton and Cole (2004) and Zaykin (2004) when HAPLO=NONE or NONEHWD.
The new RHO option in the PROC ALLELE statement requests that the LD measure 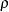 and its information
and its information 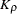 (Morton et al. 2001) be included in the "Linkage Disequilibrium Measures" table. This table now also includes a column containing the number of individuals genotyped at each pair of markers.
(Morton et al. 2001) be included in the "Linkage Disequilibrium Measures" table. This table now also includes a column containing the number of individuals genotyped at each pair of markers.
| BTL Procedure |
The experimental BTL procedure performs mixed model analysis of variance and maximum likelihood estimation on genetic marker data from experimental populations in order to find and characterize binary trait loci (BTL).
| GENESELECT Procedure |
The experimental GENESELECT procedure creates a model to predict a qualitative or quantitative phenotype from interactions of genetic and environmental variables. The procedure generates useful interactions from a potentially large number of candidates.
| References |
-
Hamilton, D.C. and Cole, D.E. (2004), "Standardizing a Composite Measure of Linkage Disequilibrium," Annals of Human Genetics, 68, 234–239.
-
Morton, N.E., Zhang,W., Taillon-Miller, P., Ennis, S., Kwok, P.-Y., and Collins A. (2001), "The Optimal Measure of Allelic Association," Proceedings of the National Academy of Sciences of the United States of America, 98, 5217–5221.
-
Zaykin, D. (2004), "Bounds and Normalization of the Composite Linkage Disequilibrium Coefficient," Genetic Epidemiology, 27, 252–257.
Copyright © 2008 by SAS Institute Inc.,Cary, NC, USA. All rights reserved.