The HPLOGISTIC Procedure
-
Overview

-
Getting Started

-
Syntax

-
Details
 Missing ValuesResponse DistributionsLog-Likelihood FunctionsExistence of Maximum Likelihood EstimatesUsing Validation and Test DataModel Fit and Assessment StatisticsThe Hosmer-Lemeshow Goodness-of-Fit TestComputational Method: MultithreadingChoosing an Optimization AlgorithmDisplayed OutputODS Table Names
Missing ValuesResponse DistributionsLog-Likelihood FunctionsExistence of Maximum Likelihood EstimatesUsing Validation and Test DataModel Fit and Assessment StatisticsThe Hosmer-Lemeshow Goodness-of-Fit TestComputational Method: MultithreadingChoosing an Optimization AlgorithmDisplayed OutputODS Table Names -
Examples

- References
The Hosmer-Lemeshow Goodness-of-Fit Test
To evaluate the fit of the model, Hosmer and Lemeshow (2000) proposed a statistic that they show, through simulation, is distributed as chi-square when there is no replication in any of the subpopulations. This goodness-of-fit test is available only for binary response models.
The unit interval is partitioned into 2,000 equal-sized bins, and each observation i is placed into the bin that contains its estimated event probability. This effectively sorts the observations in increasing order of their estimated event probability.
The observations (and frequencies) are further combined into G groups. By default G = 10, but you can specify 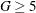 with the NGROUPS= suboption of the LACKFIT
option in the MODEL
statement. Let F be the total frequency. The target frequency for each group is
with the NGROUPS= suboption of the LACKFIT
option in the MODEL
statement. Let F be the total frequency. The target frequency for each group is 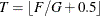 , which is the integer part of
, which is the integer part of 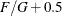 . Load the first group (
. Load the first group (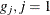 ) with the first of the 2,000 bins that has nonzero frequency
) with the first of the 2,000 bins that has nonzero frequency 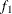 , and let the next nonzero bin have a frequency of f. PROC HPLOGISTIC performs the following steps for each nonzero bin to create the groups:
, and let the next nonzero bin have a frequency of f. PROC HPLOGISTIC performs the following steps for each nonzero bin to create the groups:
-
If
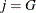 , then add this bin to group
, then add this bin to group 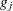 .
.
-
Otherwise, if
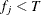 and
and 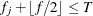 , then add this bin to group
, then add this bin to group 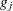 .
.
-
Otherwise, start loading the next group (
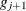 ) with
) with 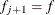 , and set
, and set 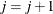 .
.
If the final group 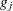 has frequency
has frequency 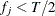 , then add these observations to the preceding group. The total number of groups actually created, g, can be less than G.
, then add these observations to the preceding group. The total number of groups actually created, g, can be less than G.
The Hosmer-Lemeshow goodness-of-fit statistic is obtained by calculating the Pearson chi-square statistic from the 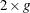 table of observed and expected frequencies. The statistic is written
table of observed and expected frequencies. The statistic is written
![\[ \chi ^2_{\mathit{HL}} = \sum _{j=1}^ g \frac{(O_ j - F_ j \bar{\pi }_ j)^2}{F_ j \bar{\pi }_ j (1 - \bar{\pi }_ j)} \]](images/statug_hplogistic0129.png)
where, for the jth group 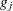 ,
, 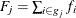 is the total frequency of subjects,
is the total frequency of subjects, 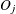 is the total frequency of event outcomes, and
is the total frequency of event outcomes, and 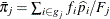 is the average estimated predicted probability of an event outcome. Let
is the average estimated predicted probability of an event outcome. Let  be the square root of the machine epsilon divided by 4,000, which is about 2.5E–12. Any
be the square root of the machine epsilon divided by 4,000, which is about 2.5E–12. Any 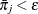 is set to
is set to  ; similarly, any
; similarly, any 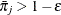 is set to
is set to 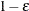 .
.
The Hosmer-Lemeshow statistic is compared to a chi-square distribution with 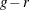 degrees of freedom. You can specify r with the DFREDUCE= suboption of the LACKFIT
option in the MODEL
statement. By default,
degrees of freedom. You can specify r with the DFREDUCE= suboption of the LACKFIT
option in the MODEL
statement. By default, 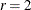 , and to compute the Hosmer-Lemeshow statistic you must have
, and to compute the Hosmer-Lemeshow statistic you must have 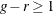 . Large values of
. Large values of 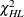 (and small p-values) indicate a lack of fit of the model.
(and small p-values) indicate a lack of fit of the model.