The MODECLUS Procedure
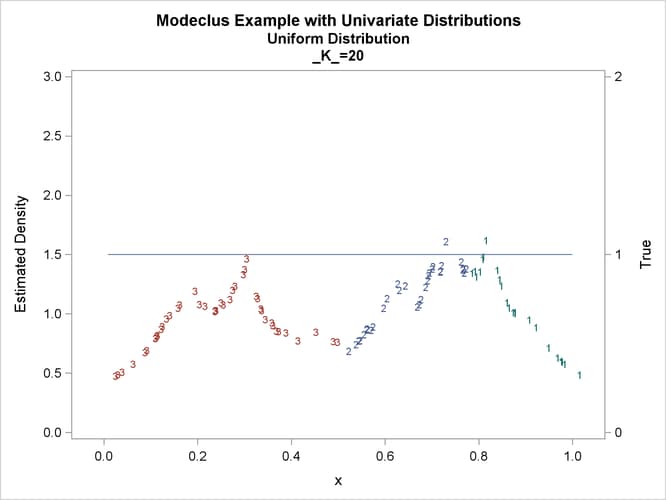
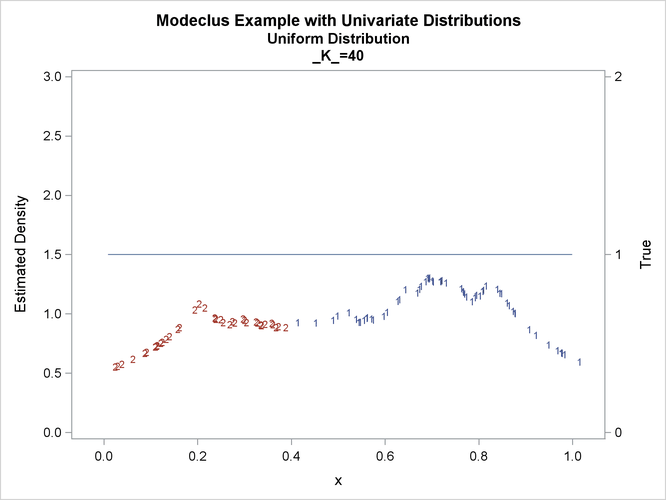
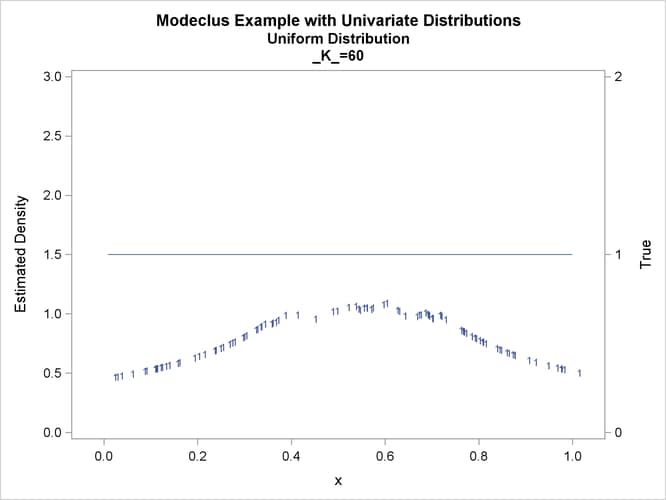
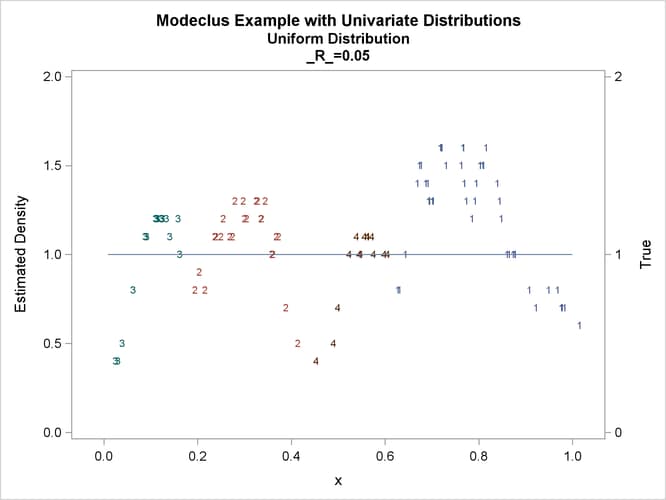
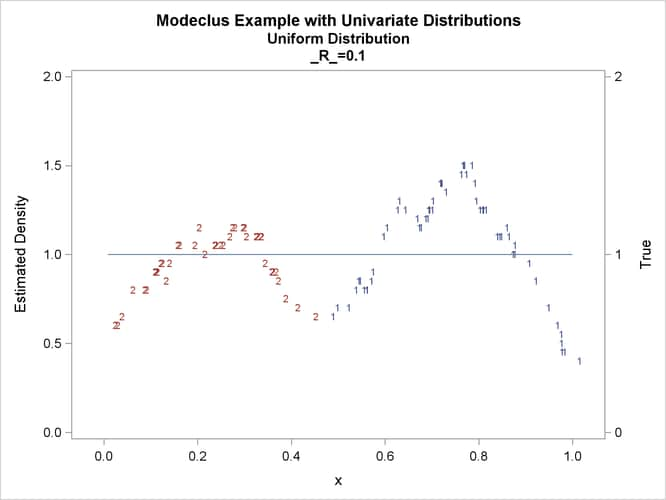
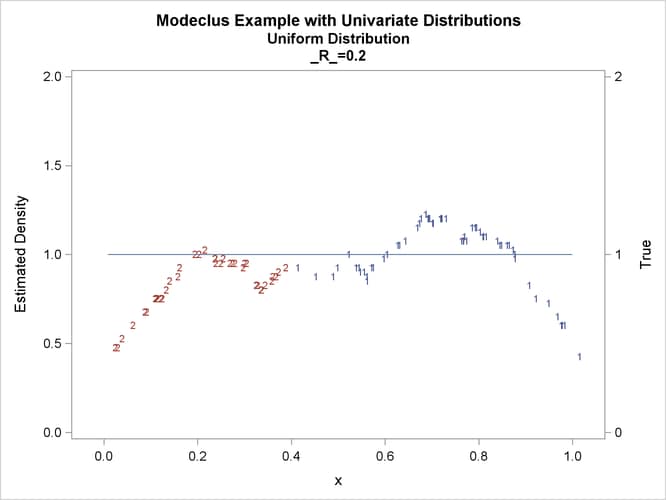
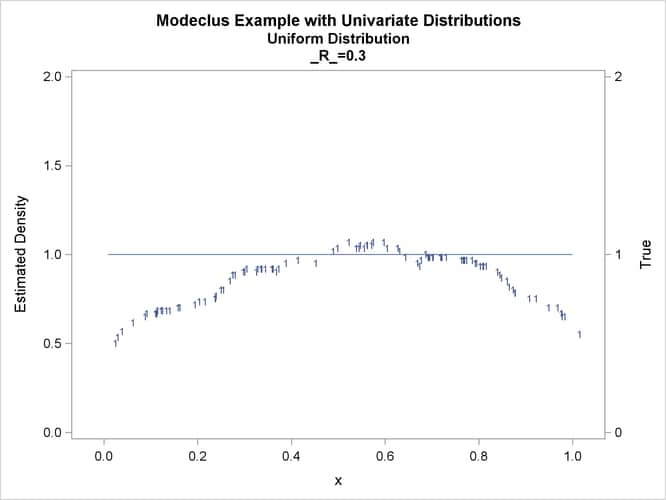
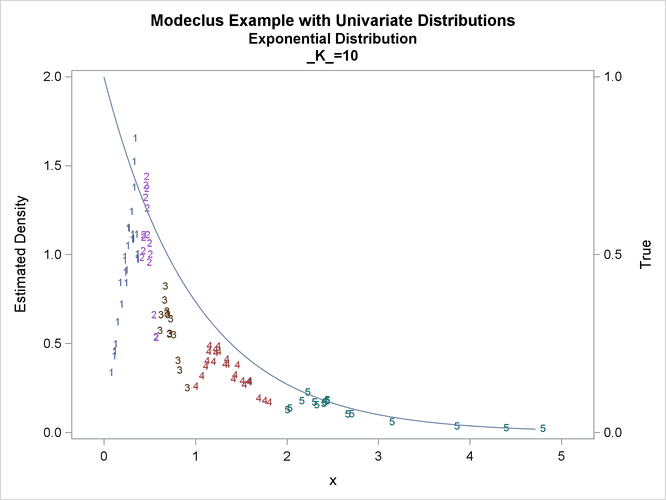
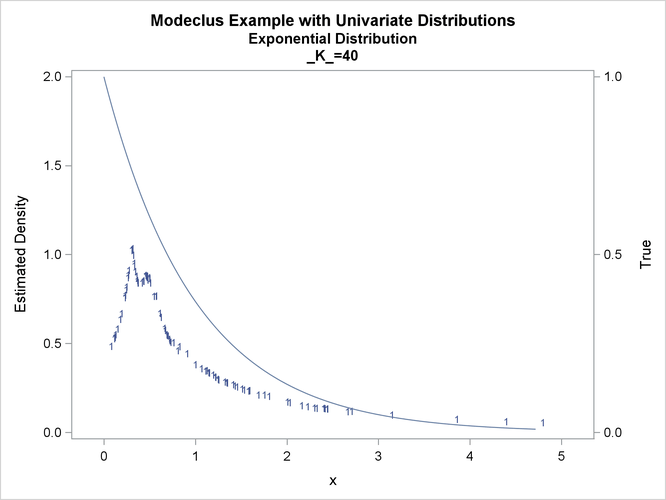
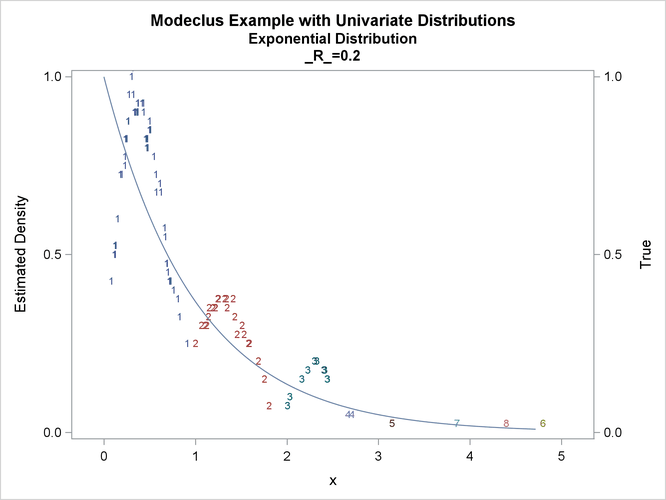
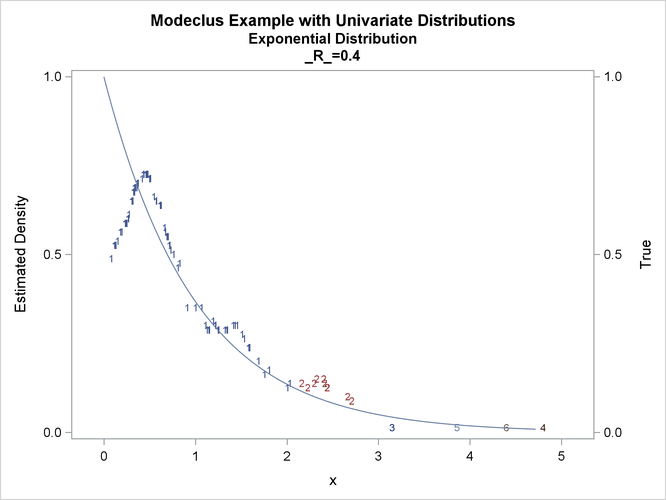
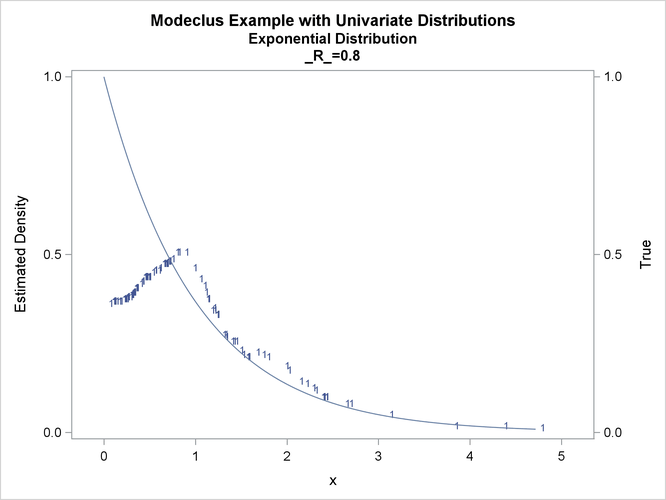
This example uses pseudo-random samples from a uniform distribution, an exponential distribution, and a bimodal mixture of two normal distributions. Results are presented in Output 66.1.1 through Output 66.1.18 as plots displaying both the true density and the estimated density, as well as cluster membership.
The following statements produce Output 66.1.1 through Output 66.1.4:
title 'Modeclus Example with Univariate Distributions';
title2 'Uniform Distribution';
data uniform;
drop n;
true=1;
do n=1 to 100;
x=ranuni(123);
output;
end;
run;
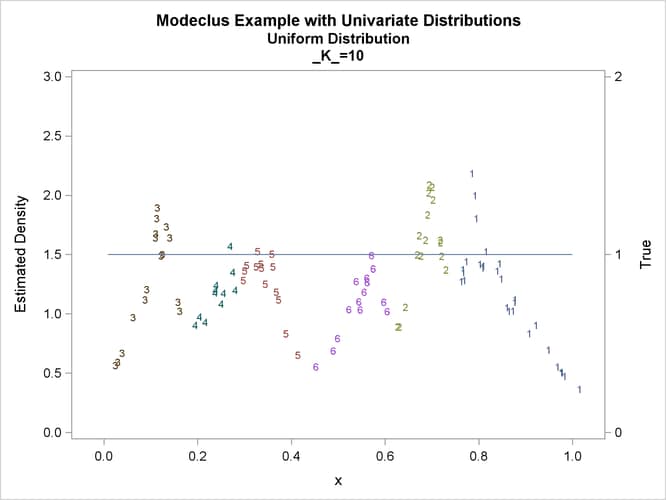
proc modeclus data=uniform m=1 k=10 20 40 60 out=out short; var x; run; proc sgplot data=out noautolegend; y2axis label='True' values=(0 to 2 by 1.); yaxis values=(0 to 3 by 0.5); scatter y=density x=x / markerchar=cluster group=cluster; pbspline y=true x=x / y2axis nomarkers lineattrs=(thickness= 1); by _K_; run;
proc modeclus data=uniform m=1 r=.05 .10 .20 .30 out=out short; var x; run; proc sgplot data=out noautolegend; y2axis label='True' values=(0 to 2 by 1.); yaxis values=(0 to 2 by 0.5); scatter y=density x=x / markerchar=cluster group=cluster; pbspline y=true x=x / y2axis nomarkers lineattrs=(thickness= 1); by _R_; run;
The following statements produce Output 66.1.5 through Output 66.1.12:
data expon;
title2 'Exponential Distribution';
drop n;
do n=1 to 100;
x=ranexp(123);
true=exp(-x);
output;
end;
run;
proc modeclus data=expon m=1 k=10 20 40 out=out short; var x; run; proc sgplot data=out noautolegend; y2axis label='True' values=(0 to 1 by .5); yaxis values=(0 to 2 by 0.5); scatter y=density x=x / markerchar=cluster group=cluster; pbspline y=true x=x / y2axis nomarkers lineattrs=(thickness= 1); by _K_; run;
proc modeclus data=expon m=1 r=.20 .40 .80 out=out short; var x; run; proc sgplot data=out noautolegend; y2axis label='True' values=(0 to 1 by .5); yaxis values=(0 to 1 by 0.5); scatter y=density x=x / markerchar=cluster group=cluster; pbspline y=true x=x / y2axis nomarkers lineattrs=(thickness= 1); by _R_; run;
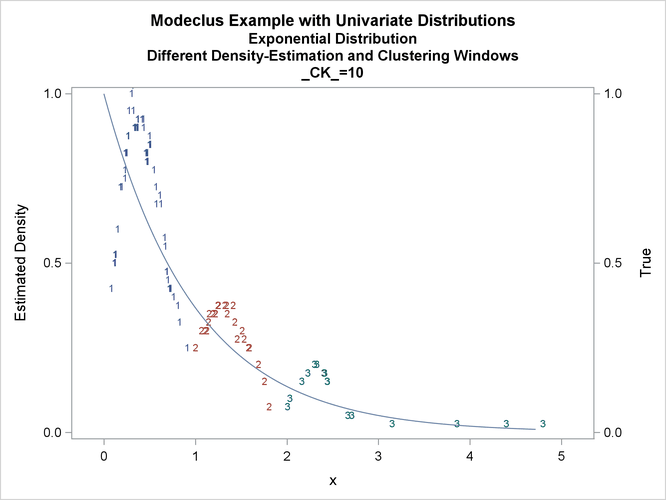
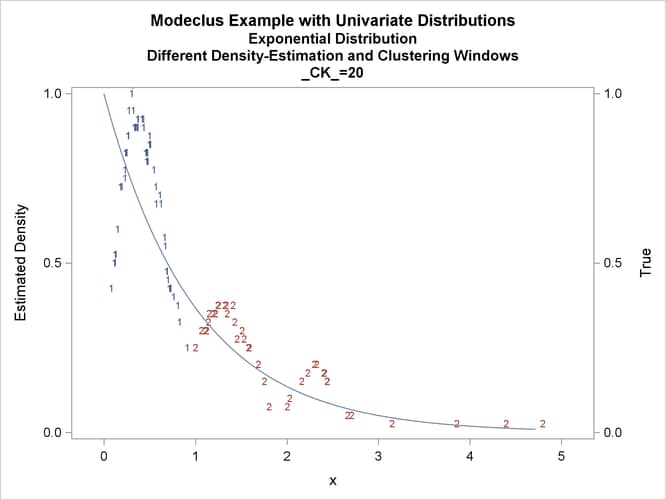
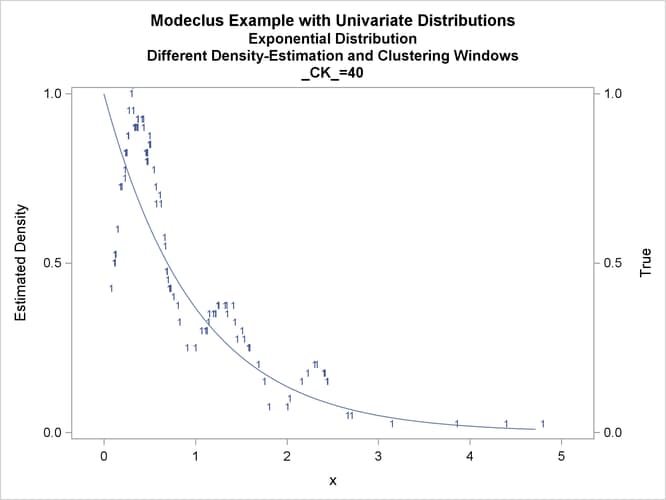
title3 'Different Density-Estimation and Clustering Windows';
proc modeclus data=expon m=1 r=.20 ck=10 20 40
out=out short;
var x;
run;
proc sgplot data=out noautolegend;
y2axis label='True' values=(0 to 1 by .5);
yaxis values=(0 to 1 by 0.5);
scatter y=density x=x / markerchar=cluster group=cluster;
pbspline y=true x=x / y2axis nomarkers lineattrs=(thickness= 1);
by _CK_;
run;
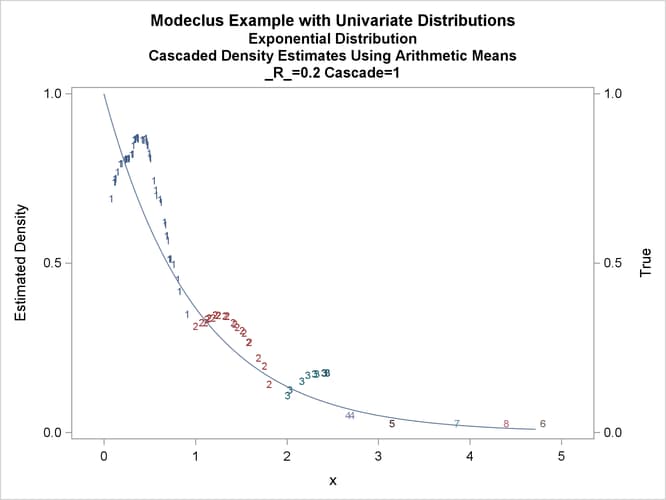
title3 'Cascaded Density Estimates Using Arithmetic Means'; proc modeclus data=expon m=1 r=.20 cascade=1 2 4 am out=out short; var x; run; proc sgplot data=out noautolegend; y2axis label='True' values=(0 to 1 by .5); yaxis values=(0 to 1 by 0.5); scatter y=density x=x / markerchar=cluster group=cluster; pbspline y=true x=x / y2axis nomarkers lineattrs=(thickness= 1); by _R_ _CASCAD_; run;
Output 66.1.10: True Density, Estimated Density, and Cluster Membership by _R_=0.2 with Various _CK_ Values
Output 66.1.12: True Density, Estimated Density, and Cluster Membership by _R_=0.2 with Various _CASCAD_ Values
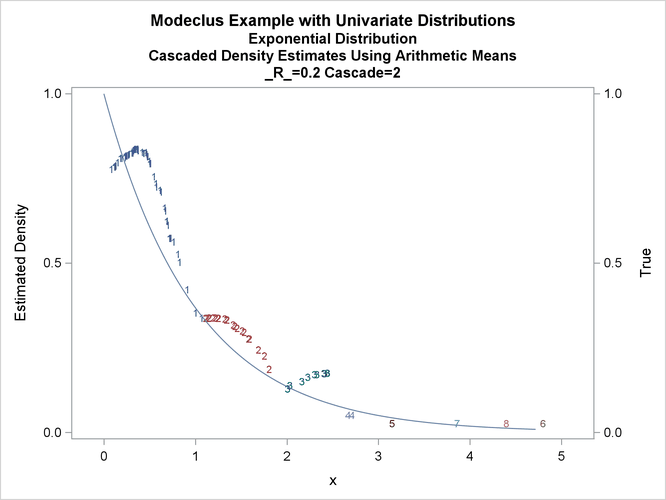
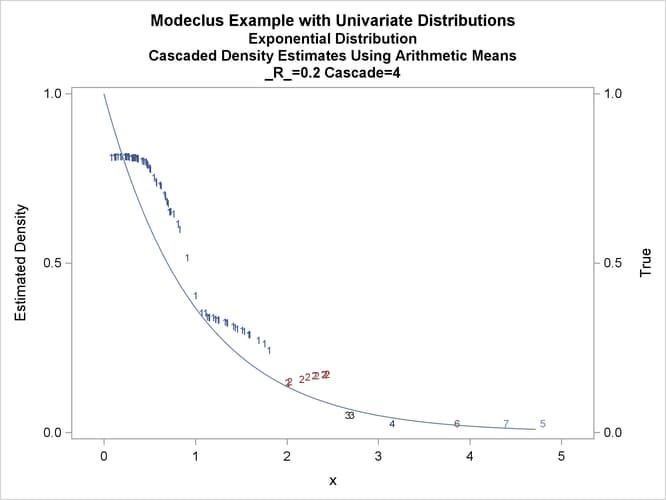
The following statements produce Output 66.1.13 through Output 66.1.18:
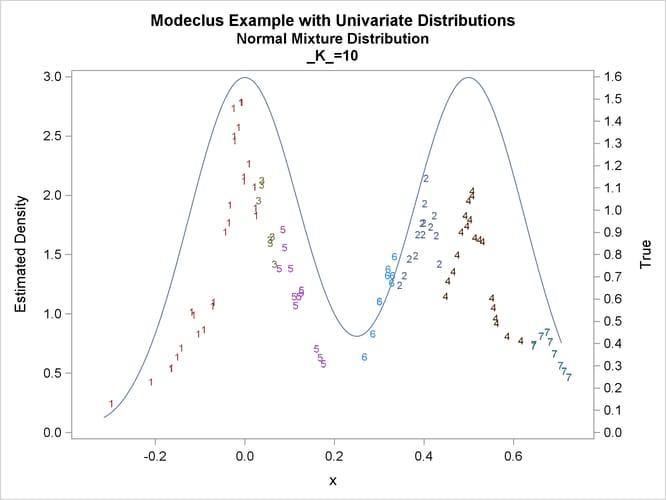
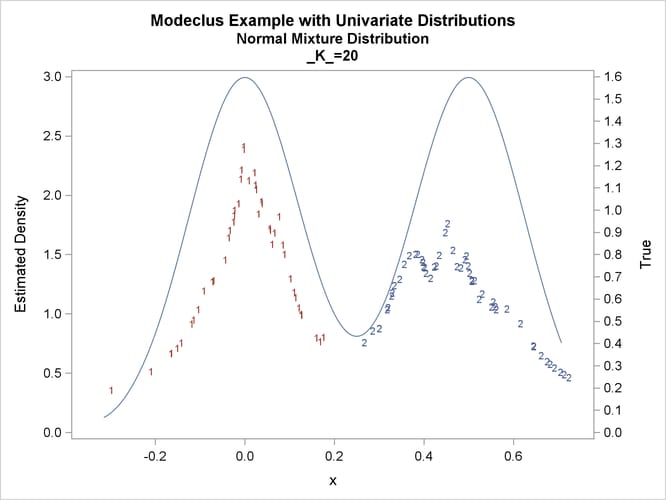
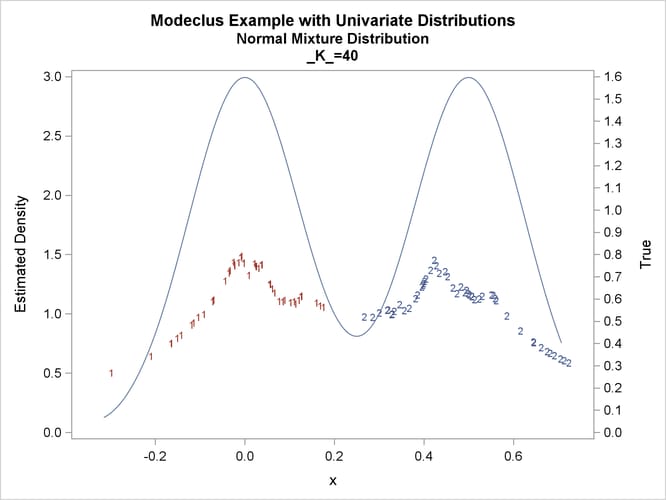
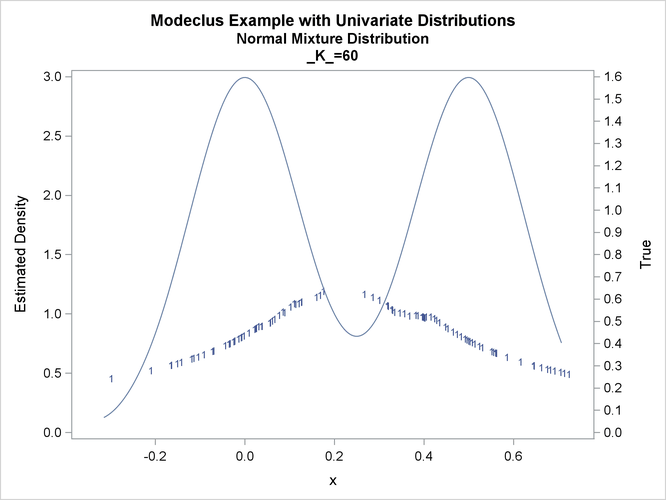
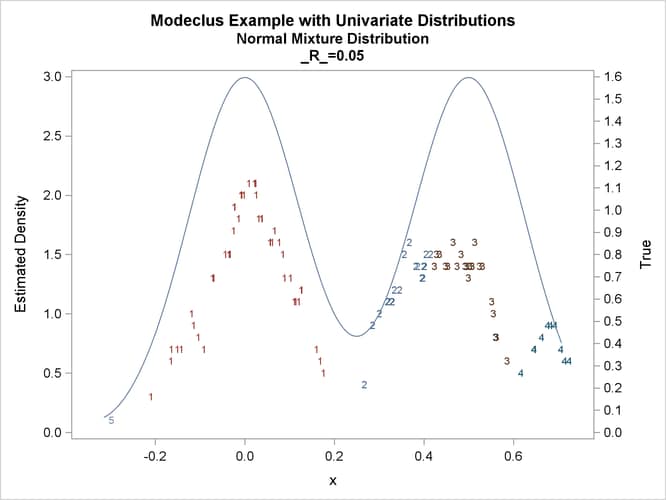
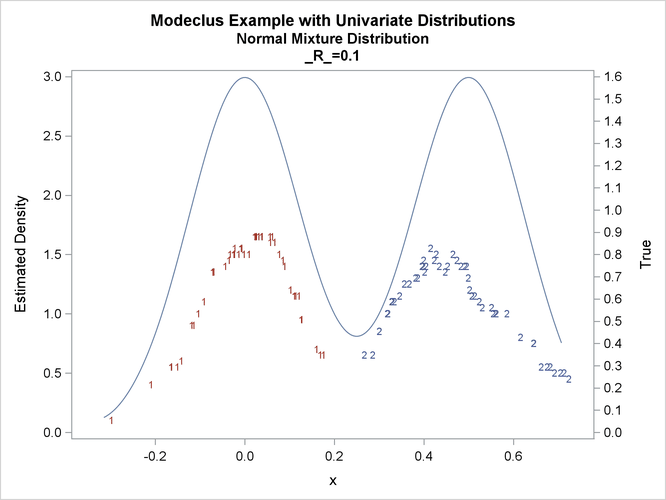
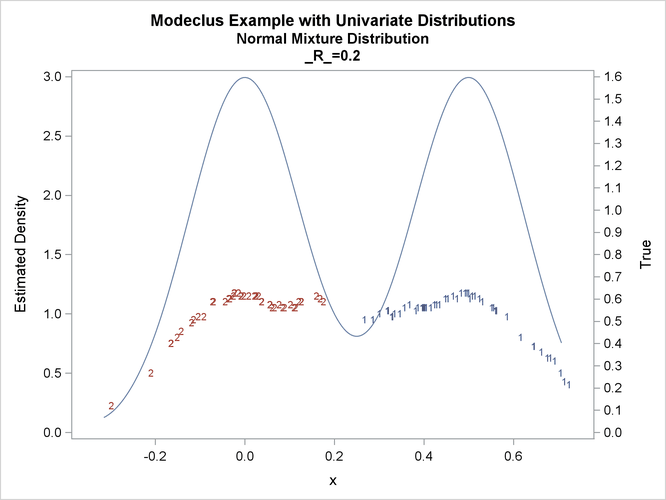
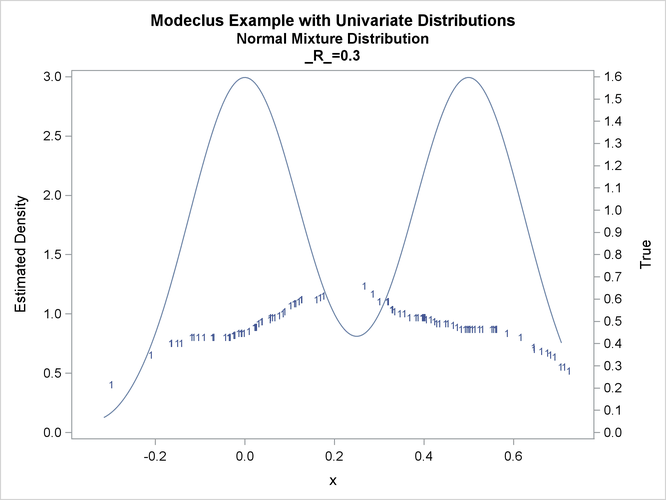
title2 'Normal Mixture Distribution';
data normix;
drop n sigma;
sigma=.125;
do n=1 to 100;
x=rannor(456)*sigma+mod(n,2)/2;
true=exp(-.5*(x/sigma)**2)+exp(-.5*((x-.5)/sigma)**2);
true=.5*true/(sigma*sqrt(2*3.1415926536));
output;
end;
run;
proc modeclus data=normix m=1 k=10 20 40 60 out=out short; var x; run; proc sgplot data=out noautolegend; y2axis label='True' values=(0 to 1.6 by .1); yaxis values=(0 to 3 by 0.5); scatter y=density x=x / markerchar=cluster group=cluster; pbspline y=true x=x / y2axis nomarkers lineattrs=(thickness= 1); by _K_; run;
proc modeclus data=normix m=1 r=.05 .10 .20 .30 out=out short; var x; run; proc sgplot data=out noautolegend; y2axis label='True' values=(0 to 1.6 by .1); yaxis values=(0 to 3 by 0.5); scatter y=density x=x / markerchar=cluster group=cluster; pbspline y=true x=x / y2axis nomarkers lineattrs=(thickness= 1); by _R_; run;
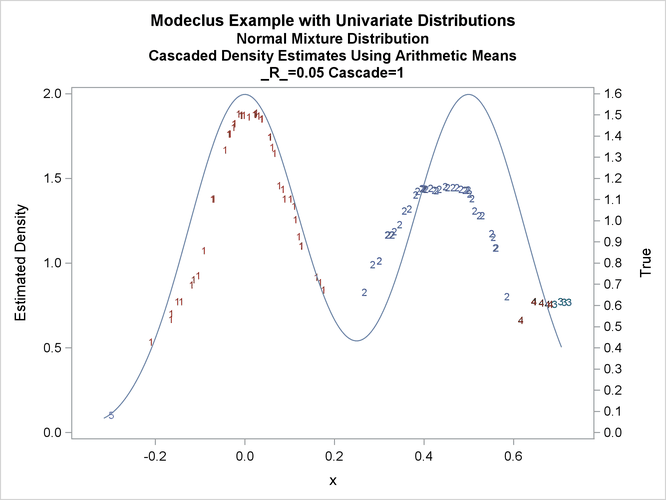
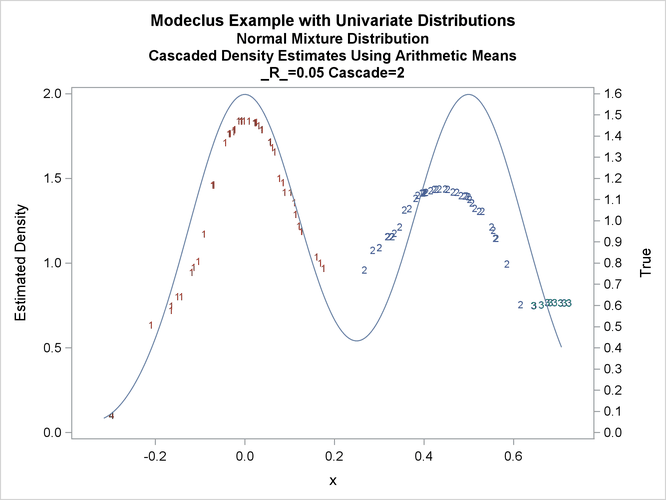
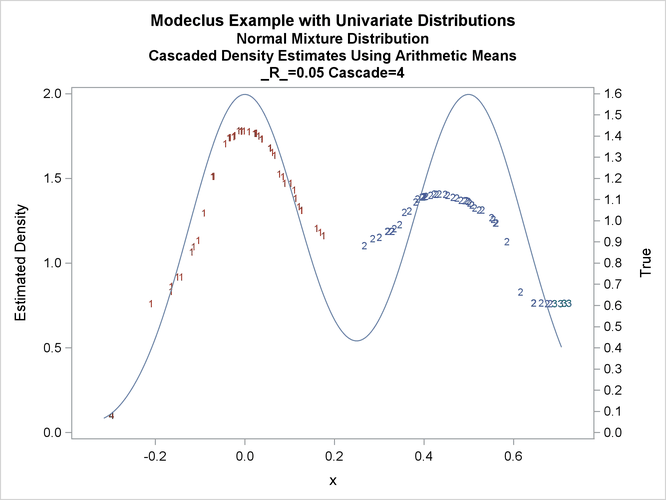
title3 'Cascaded Density Estimates Using Arithmetic Means'; proc modeclus data=normix m=1 r=.05 cascade=1 2 4 am out=out short; var x; run; proc sgplot data=out noautolegend; y2axis label='True' values=(0 to 1.6 by .1); yaxis values=(0 to 2 by 0.5); scatter y=density x=x / markerchar=cluster group=cluster; pbspline y=true x=x / y2axis nomarkers lineattrs=(thickness= 1); by _R_ _CASCAD_; run;
Output 66.1.18: True Density, Estimated Density, and Cluster Membership by _R_=0.05 with Various _CASCAD_ Values