The SURVEYFREQ Procedure
The Jackknife Method
If you specify the VARMETHOD=JACKKNIFE option, PROC SURVEYFREQ uses the delete-1 jackknife method for variance estimation. The jackknife method can be used for stratified sample designs and for designs with no stratification. If your design is stratified, the jackknife method requires at least two PSUs in each stratum. You can provide replicate weights for jackknife variance estimation by using a REPWEIGHTS statement, or the procedure can construct replicate weights for the analysis. PROC SURVEYFREQ estimates the parameter of interest (a proportion, total, odds ratio, or other statistic) from each replicate, and then uses the variability among replicate estimates to estimate the overall variance of the parameter estimate. See Wolter (1985) and Lohr (2009) for more information about jackknife variance estimation.
If you do not provide replicate weights with a REPWEIGHTS statement, PROC SURVEYFREQ constructs the replicates. The number of replicates  equals the number of PSUs, and the procedure deletes one PSU from the full sample to form each replicate. The sampling weights are modified by the jackknife coefficient for the replicate to create the replicate weights.
equals the number of PSUs, and the procedure deletes one PSU from the full sample to form each replicate. The sampling weights are modified by the jackknife coefficient for the replicate to create the replicate weights.
If your design is not stratified (no STRATA statement), the jackknife coefficient has the same value for each replicate  . The jackknife coefficient equals
. The jackknife coefficient equals
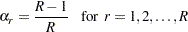 |
where  is the total number of replicates (or total number of PSUs). For the PSUs included in a replicate, the replicate weights are computed by dividing the original sampling weights by the jackknife coefficient. For the deleted PSU, which is not included in the replicate, the replicate weights equal zero. The replicate weight for the
is the total number of replicates (or total number of PSUs). For the PSUs included in a replicate, the replicate weights are computed by dividing the original sampling weights by the jackknife coefficient. For the deleted PSU, which is not included in the replicate, the replicate weights equal zero. The replicate weight for the  th member of the
th member of the  th PSU can be expressed as follows when the design is not stratified:
th PSU can be expressed as follows when the design is not stratified:
 |
where  is the original sampling weight of unit
is the original sampling weight of unit  ,
,  is the replicate number, and
is the replicate number, and 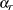 is the jackknife coefficient.
is the jackknife coefficient.
If your design is stratified, the jackknife method requires at least two PSUs in each stratum. Let stratum 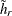 be the stratum from which a PSU is deleted to form the
be the stratum from which a PSU is deleted to form the  th replicate. Stratum
th replicate. Stratum 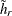 is called the donor stratum. The jackknife coefficients are defined as
is called the donor stratum. The jackknife coefficients are defined as
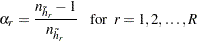 |
where 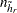 is the total number of PSUs in the donor stratum for replicate
is the total number of PSUs in the donor stratum for replicate  . For all strata other than the donor stratum, the replicate
. For all strata other than the donor stratum, the replicate  weights equal the original sampling weights. For PSUs included from the donor stratum, the replicate weights are computed by dividing the original sampling weights by the jackknife coefficient. For the deleted PSU, which is not included in the replicate, the replicate weights equal zero. The replicate weight for the
weights equal the original sampling weights. For PSUs included from the donor stratum, the replicate weights are computed by dividing the original sampling weights by the jackknife coefficient. For the deleted PSU, which is not included in the replicate, the replicate weights equal zero. The replicate weight for the  th member of the
th member of the 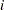 th PSU in stratum
th PSU in stratum  can be expressed as
can be expressed as
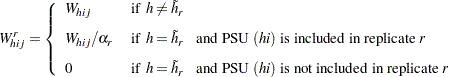 |
You can use the OUTWEIGHTS= method-option to store the replicate weights in a SAS data set. You can also use the OUTJKCOEFS= method-option to store the jackknife coefficients in a SAS data set. See the sections Jackknife Coefficients Output Data Set and Replicate Weights Output Data Set for details about the contents of these output data sets. You can provide replicate weights and jackknife coefficients to the procedure for subsequent analyses by using a REPWEIGHTS statement. If you provide replicate weights but do not provide jackknife coefficients, PROC SURVEYFREQ uses 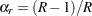 as the jackknife coefficient for all replicates.
as the jackknife coefficient for all replicates.
Let 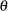 denote the population parameter to be estimated—for example, a proportion, total, odds ratio, or other statistic. Let
denote the population parameter to be estimated—for example, a proportion, total, odds ratio, or other statistic. Let 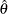 denote the estimate of
denote the estimate of 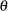 from the full sample, and let
from the full sample, and let 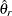 be the estimate from the
be the estimate from the  th jackknife replicate, which is computed by using the replicate weights. The jackknife variance estimate for
th jackknife replicate, which is computed by using the replicate weights. The jackknife variance estimate for 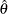 is computed as
is computed as
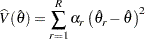 |
where  is the total number of replicates and
is the total number of replicates and 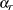 is the jackknife coefficient for replicate
is the jackknife coefficient for replicate  .
.
If a parameter cannot be estimated from some replicate(s), then the variance estimate is computed by using those replicates from which the parameter can be estimated. For example, suppose the parameter is a column proportion—the proportion of column  for table cell
for table cell 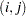 . If a replicate
. If a replicate  contains no observations in column
contains no observations in column  , then the column
, then the column  proportion is not estimable from replicate
proportion is not estimable from replicate  . In this case, the jackknife variance estimate is computed as
. In this case, the jackknife variance estimate is computed as
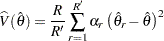 |
where the summation is over the replicates where the parameter 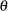 is estimable, and
is estimable, and  is the number of those replicates.
is the number of those replicates.
