The LIFETEST Procedure
Example 51.2 Enhanced Survival Plot and Multiple-Comparison Adjustments
This example highlights a number of features in the survival plot that uses ODS Graphics. Also shown in this example are comparisons of survival curves based on multiple comparison adjustments. Data of 137 bone marrow transplant patients extracted from Klein and Moeschberger (1997) have been saved in the data set BMT in the Sashelp library. At the time of transplant, each patient is classified into one of three risk categories: ALL (acute lymphoblastic leukemia), AML (acute myelocytic leukemia)-Low Risk, and AML-High Risk. The endpoint of interest is the disease-free survival time, which is the time to death or relapse or to the end of the study in days. In this data set, the variable Group represents the patient’s risk category, the variable T represents the disease-free survival time, and the variable Status is the censoring indicator, with the value 1 indicating an event time and the value 0 a censored time.
The following step displays the first 10 observations of the BMT data set in Output 51.2.1. The data set is available in the Sashelp library.
proc print data=Sashelp.BMT(obs=10); run;
| Obs | Group | T | Status |
|---|---|---|---|
| 1 | ALL | 2081 | 0 |
| 2 | ALL | 1602 | 0 |
| 3 | ALL | 1496 | 0 |
| 4 | ALL | 1462 | 0 |
| 5 | ALL | 1433 | 0 |
| 6 | ALL | 1377 | 0 |
| 7 | ALL | 1330 | 0 |
| 8 | ALL | 996 | 0 |
| 9 | ALL | 226 | 0 |
| 10 | ALL | 1199 | 0 |
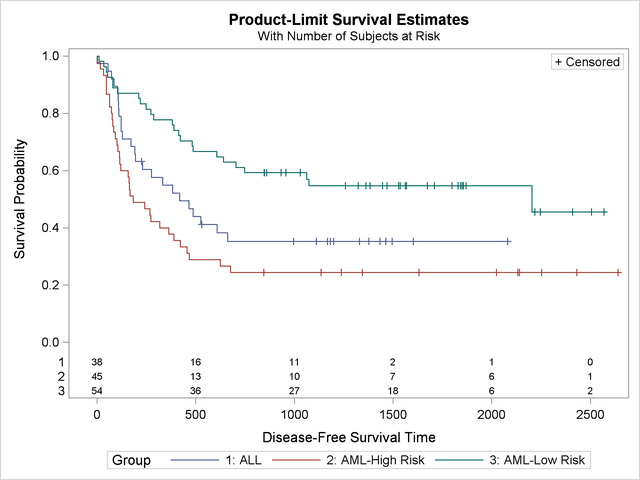
In the following statements, PROC LIFETEST is invoked to compute the product-limit estimate of the survivor function for each risk category. Using ODS Graphics, you can display the number of subjects at risk in the survival plot. The PLOTS= option requests that the survival curves be plotted, and the ATRISK= suboption specifies the time points at which the at-risk numbers are displayed. In the STRATA statement, the ADJUST=SIDAK option requests the Šidák multiple-comparison adjustment, and by default, all paired comparisons are carried out.
ods graphics on; proc lifetest data=sashelp.BMT plots=survival(atrisk=0 to 2500 by 500); time T * Status(0); strata Group / test=logrank adjust=sidak; run;
Output 51.2.2 displays the estimated disease-free survival for the three leukemia groups with the number of subjects at risk at 0, 500, 1,000, 1,500, 2,000, and 2,500 days. Patients in the AML-Low Risk group experience a longer disease-free survival than those in the ALL group, who in turn fare better than those in the AML-High Risk group.
The log-rank test (Output 51.2.3) shows that the disease-free survival times for these three risk groups are significantly different (p=0.001).
| Test of Equality over Strata | |||
|---|---|---|---|
| Test | Chi-Square | DF | Pr > Chi-Square |
| Log-Rank | 13.8037 | 2 | 0.0010 |
The  idák multiple-comparison results are shown in Output 51.2.4. There is no significant difference in disease-free survivor functions between the ALL and AML-High Risk groups (p=0.2779). The difference between the ALL and AML-Low Risk groups is marginal (p=0.0685), but the AML-Low Risk and AML-High Risk groups have significantly different disease-free survivor functions (p=0.0006).
idák multiple-comparison results are shown in Output 51.2.4. There is no significant difference in disease-free survivor functions between the ALL and AML-High Risk groups (p=0.2779). The difference between the ALL and AML-Low Risk groups is marginal (p=0.0685), but the AML-Low Risk and AML-High Risk groups have significantly different disease-free survivor functions (p=0.0006).
| Adjustment for Multiple Comparisons for the Logrank Test | ||||
|---|---|---|---|---|
| Strata Comparison | Chi-Square | p-Values | ||
| Group | Group | Raw | Sidak | |
| ALL | AML-High Risk | 2.6610 | 0.1028 | 0.2779 |
| ALL | AML-Low Risk | 5.1400 | 0.0234 | 0.0685 |
| AML-High Risk | AML-Low Risk | 13.8011 | 0.0002 | 0.0006 |
Suppose you consider the AML-Low Risk group as the reference group. You can use the DIFF= option in the STRATA statement to designate this risk group as the control and apply a multiple-comparison adjustment to the p-values for the paired comparison between the AML-Low Risk group with each of the other groups. Consider the  idák correction again. You specify the ADJUST= and DIFF= options as in the following statements:
idák correction again. You specify the ADJUST= and DIFF= options as in the following statements:
proc lifetest data=sashelp.BMT notable plots=none;
time T * Status(0);
strata Group / test=logrank adjust=sidak diff=control('AML-Low Risk');
run;
Output 51.2.5 shows that although both the ALL and AML-High Risk groups differ from the AML-Low Risk group at the 0.05 level, the difference between the AML-High Risk and the AML-Low Risk group is highly significant (p=0.0004).
| Adjustment for Multiple Comparisons for the Logrank Test | ||||
|---|---|---|---|---|
| Strata Comparison | Chi-Square | p-Values | ||
| Group | Group | Raw | Sidak | |
| ALL | AML-Low Risk | 5.1400 | 0.0234 | 0.0462 |
| AML-High Risk | AML-Low Risk | 13.8011 | 0.0002 | 0.0004 |
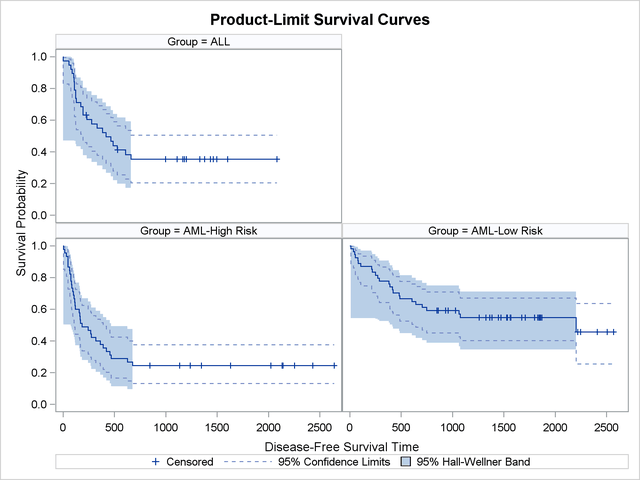
Klein and Moeschberger (1997, Section 4.4) describe in detail how to compute the Hall-Wellner (HW) and equal-precision (EP) confidence bands for the survivor function. You can output these simultaneous confidence intervals to a SAS data set by using the CONFBAND= and OUTSURV= options in the PROC LIFETEST statement. You can display survival curves with pointwise and simultaneous confidence limits through ODS Graphics. When the survival data are stratified, displaying all the survival curves and their confidence limits in the same plot can make the plot appear cluttered. In the following statements, the PLOTS= specification requests that the survivor functions be displayed along with their pointwise confidence limits (CL) and Hall-Wellner confidence bands (CB=HW). The STRATA=PANEL specification requests that the survival curves be displayed in a panel of three plots, one for each risk group.
proc lifetest data=sashelp.BMT plots=survival(cl cb=hw strata=panel); time T * Status(0); strata Group; run; ods graphics off;
The panel plot is shown in Output 51.2.6.