| The PROBIT Procedure |
| Displayed Output |
If you request the iteration history (ITPRINT), PROC PROBIT displays the following:
the current value of the log likelihood
the ridging parameter for the modified Newton-Raphson optimization process
the current estimate of the parameters
the current estimate of the parameter
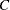 for a natural (threshold) model
for a natural (threshold) model the values of the gradient and the Hessian on the last iteration
If you include classification variables, PROC PROBIT displays the following:
the numbers of levels for each classification variable
the (ordered) values of the levels
the number of observations used
After the model is fit, PROC PROBIT displays the following:
the name of the input data set
the name of the dependent variables
the number of observations used
the number of events and the number of trials
the final value of the log-likelihood function
the parameter estimates
the standard error estimates of the parameter estimates
approximate chi-square test statistics for the test
If you specify the COVB or CORRB options, PROC PROBIT displays the following:
the estimated covariance matrix for the parameter estimates
the estimated correlation matrix for the parameter estimates
If you specify the LACKFIT option, PROC PROBIT displays the following:
a count of the number of levels of the response and the number of distinct sets of independent variables
a goodness-of-fit test based on the Pearson’s chi-square
a goodness-of-fit test based on the likelihood-ratio chi-square
If you specify only one independent variable, the normal distribution is used to model the probabilities, and the response is binary, then PROC PROBIT displays the following:
the mean MU of the stimulus tolerance
the scale parameter SIGMA of the stimulus tolerance
the covariance matrix for MU, SIGMA, and the natural response parameter
If you specify the INVERSECL options, PROC PROBIT also displays the following:
the estimated dose along with the 95% fiducial limits for probability levels 0.01 to 0.10, 0.15 to 0.85 by 0.05, and 0.90 to 0.99
Copyright © SAS Institute, Inc. All Rights Reserved.