| The LOGISTIC Procedure |
Example 51.1 Stepwise Logistic Regression and Predicted Values
Consider a study on cancer remission (Lee; 1974). The data consist of patient characteristics and whether or not cancer remission occured. The following DATA step creates the data set Remission containing seven variables. The variable remiss is the cancer remission indicator variable with a value of 1 for remission and a value of 0 for nonremission. The other six variables are the risk factors thought to be related to cancer remission.
data Remission; input remiss cell smear infil li blast temp; label remiss='Complete Remission'; datalines; 1 .8 .83 .66 1.9 1.1 .996 1 .9 .36 .32 1.4 .74 .992 0 .8 .88 .7 .8 .176 .982 0 1 .87 .87 .7 1.053 .986 1 .9 .75 .68 1.3 .519 .98 0 1 .65 .65 .6 .519 .982 1 .95 .97 .92 1 1.23 .992 0 .95 .87 .83 1.9 1.354 1.02 0 1 .45 .45 .8 .322 .999 0 .95 .36 .34 .5 0 1.038 0 .85 .39 .33 .7 .279 .988 0 .7 .76 .53 1.2 .146 .982 0 .8 .46 .37 .4 .38 1.006 0 .2 .39 .08 .8 .114 .99 0 1 .9 .9 1.1 1.037 .99 1 1 .84 .84 1.9 2.064 1.02 0 .65 .42 .27 .5 .114 1.014 0 1 .75 .75 1 1.322 1.004 0 .5 .44 .22 .6 .114 .99 1 1 .63 .63 1.1 1.072 .986 0 1 .33 .33 .4 .176 1.01 0 .9 .93 .84 .6 1.591 1.02 1 1 .58 .58 1 .531 1.002 0 .95 .32 .3 1.6 .886 .988 1 1 .6 .6 1.7 .964 .99 1 1 .69 .69 .9 .398 .986 0 1 .73 .73 .7 .398 .986 ;
The following invocation of PROC LOGISTIC illustrates the use of stepwise selection to identify the prognostic factors for cancer remission. A significance level of 0.3 is required to allow a variable into the model (SLENTRY=0.3), and a significance level of 0.35 is required for a variable to stay in the model (SLSTAY=0.35). A detailed account of the variable selection process is requested by specifying the DETAILS option. The Hosmer and Lemeshow goodness-of-fit test for the final selected model is requested by specifying the LACKFIT option. The OUTEST= and COVOUT options in the PROC LOGISTIC statement create a data set that contains parameter estimates and their covariances for the final selected model. The response variable option EVENT= chooses remiss=1 (remission) as the event so that the probability of remission is modeled. The OUTPUT statement creates a data set that contains the cumulative predicted probabilities and the corresponding confidence limits, and the individual and cross validated predicted probabilities for each observation.
title 'Stepwise Regression on Cancer Remission Data';
proc logistic data=Remission outest=betas covout;
model remiss(event='1')=cell smear infil li blast temp
/ selection=stepwise
slentry=0.3
slstay=0.35
details
lackfit;
output out=pred p=phat lower=lcl upper=ucl
predprob=(individual crossvalidate);
run;
proc print data=betas; title2 'Parameter Estimates and Covariance Matrix'; run;
proc print data=pred; title2 'Predicted Probabilities and 95% Confidence Limits'; run;
In stepwise selection, an attempt is made to remove any insignificant variables from the model before adding a significant variable to the model. Each addition or deletion of a variable to or from a model is listed as a separate step in the displayed output, and at each step a new model is fitted. Details of the model selection steps are shown in Outputs 51.1.1 through 51.1.5.
Prior to the first step, the intercept-only model is fit and individual score statistics for the potential variables are evaluated (Output 51.1.1).
| Stepwise Regression on Cancer Remission Data |
| Model Convergence Status |
|---|
| Convergence criterion (GCONV=1E-8) satisfied. |
In Step 1 (Output 51.1.2), the variable li is selected into the model since it is the most significant variable among those to be chosen (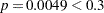 ). The intermediate model that contains an intercept and li is then fitted. li remains significant (
). The intermediate model that contains an intercept and li is then fitted. li remains significant (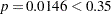 ) and is not removed.
) and is not removed.
| Model Fit Statistics | ||
|---|---|---|
| Criterion | Intercept Only |
Intercept and Covariates |
| AIC | 36.372 | 30.073 |
| SC | 37.668 | 32.665 |
| -2 Log L | 34.372 | 26.073 |
| Testing Global Null Hypothesis: BETA=0 | |||
|---|---|---|---|
| Test | Chi-Square | DF | Pr > ChiSq |
| Likelihood Ratio | 8.2988 | 1 | 0.0040 |
| Score | 7.9311 | 1 | 0.0049 |
| Wald | 5.9594 | 1 | 0.0146 |
| Analysis of Maximum Likelihood Estimates | |||||
|---|---|---|---|---|---|
| Parameter | DF | Estimate | Standard Error |
Wald Chi-Square |
Pr > ChiSq |
| Intercept | 1 | -3.7771 | 1.3786 | 7.5064 | 0.0061 |
| li | 1 | 2.8973 | 1.1868 | 5.9594 | 0.0146 |
In Step 2 (Output 51.1.3), the variable temp is added to the model. The model then contains an intercept and the variables li and temp. Both li and temp remain significant at 0.35 level; therefore, neither li nor temp is removed from the model.
| Model Fit Statistics | ||
|---|---|---|
| Criterion | Intercept Only |
Intercept and Covariates |
| AIC | 36.372 | 30.648 |
| SC | 37.668 | 34.535 |
| -2 Log L | 34.372 | 24.648 |
| Testing Global Null Hypothesis: BETA=0 | |||
|---|---|---|---|
| Test | Chi-Square | DF | Pr > ChiSq |
| Likelihood Ratio | 9.7239 | 2 | 0.0077 |
| Score | 8.3648 | 2 | 0.0153 |
| Wald | 5.9052 | 2 | 0.0522 |
| Analysis of Maximum Likelihood Estimates | |||||
|---|---|---|---|---|---|
| Parameter | DF | Estimate | Standard Error |
Wald Chi-Square |
Pr > ChiSq |
| Intercept | 1 | 47.8448 | 46.4381 | 1.0615 | 0.3029 |
| li | 1 | 3.3017 | 1.3593 | 5.9002 | 0.0151 |
| temp | 1 | -52.4214 | 47.4897 | 1.2185 | 0.2697 |
| Odds Ratio Estimates | |||
|---|---|---|---|
| Effect | Point Estimate | 95% Wald Confidence Limits |
|
| li | 27.158 | 1.892 | 389.856 |
| temp | <0.001 | <0.001 | >999.999 |
| Association of Predicted Probabilities and Observed Responses |
|||
|---|---|---|---|
| Percent Concordant | 87.0 | Somers' D | 0.747 |
| Percent Discordant | 12.3 | Gamma | 0.752 |
| Percent Tied | 0.6 | Tau-a | 0.345 |
| Pairs | 162 | c | 0.873 |
In Step 3 (Output 51.1.4), the variable cell is added to the model. The model then contains an intercept and the variables li, temp, and cell. None of these variables are removed from the model since all are significant at the 0.35 level.
| Model Fit Statistics | ||
|---|---|---|
| Criterion | Intercept Only |
Intercept and Covariates |
| AIC | 36.372 | 29.953 |
| SC | 37.668 | 35.137 |
| -2 Log L | 34.372 | 21.953 |
| Testing Global Null Hypothesis: BETA=0 | |||
|---|---|---|---|
| Test | Chi-Square | DF | Pr > ChiSq |
| Likelihood Ratio | 12.4184 | 3 | 0.0061 |
| Score | 9.2502 | 3 | 0.0261 |
| Wald | 4.8281 | 3 | 0.1848 |
| Analysis of Maximum Likelihood Estimates | |||||
|---|---|---|---|---|---|
| Parameter | DF | Estimate | Standard Error |
Wald Chi-Square |
Pr > ChiSq |
| Intercept | 1 | 67.6339 | 56.8875 | 1.4135 | 0.2345 |
| cell | 1 | 9.6521 | 7.7511 | 1.5507 | 0.2130 |
| li | 1 | 3.8671 | 1.7783 | 4.7290 | 0.0297 |
| temp | 1 | -82.0737 | 61.7124 | 1.7687 | 0.1835 |
| Odds Ratio Estimates | |||
|---|---|---|---|
| Effect | Point Estimate | 95% Wald Confidence Limits |
|
| cell | >999.999 | 0.004 | >999.999 |
| li | 47.804 | 1.465 | >999.999 |
| temp | <0.001 | <0.001 | >999.999 |
| Association of Predicted Probabilities and Observed Responses |
|||
|---|---|---|---|
| Percent Concordant | 88.9 | Somers' D | 0.778 |
| Percent Discordant | 11.1 | Gamma | 0.778 |
| Percent Tied | 0.0 | Tau-a | 0.359 |
| Pairs | 162 | c | 0.889 |
Finally, none of the remaining variables outside the model meet the entry criterion, and the stepwise selection is terminated. A summary of the stepwise selection is displayed in Output 51.1.5.
Results of the Hosmer and Lemeshow test are shown in Output 51.1.6. There is no evidence of a lack of fit in the selected model 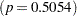 .
.
The data set betas created by the OUTEST= and COVOUT options is displayed in Output 51.1.7. The data set contains parameter estimates and the covariance matrix for the final selected model. Note that all explanatory variables listed in the MODEL statement are included in this data set; however, variables that are not included in the final model have all missing values.
| Stepwise Regression on Cancer Remission Data |
| Parameter Estimates and Covariance Matrix |
| Obs | _LINK_ | _TYPE_ | _STATUS_ | _NAME_ | Intercept | cell | smear | infil | li | blast | temp | _LNLIKE_ |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | LOGIT | PARMS | 0 Converged | remiss | 67.63 | 9.652 | . | . | 3.8671 | . | -82.07 | -10.9767 |
| 2 | LOGIT | COV | 0 Converged | Intercept | 3236.19 | 157.097 | . | . | 64.5726 | . | -3483.23 | -10.9767 |
| 3 | LOGIT | COV | 0 Converged | cell | 157.10 | 60.079 | . | . | 6.9454 | . | -223.67 | -10.9767 |
| 4 | LOGIT | COV | 0 Converged | smear | . | . | . | . | . | . | . | -10.9767 |
| 5 | LOGIT | COV | 0 Converged | infil | . | . | . | . | . | . | . | -10.9767 |
| 6 | LOGIT | COV | 0 Converged | li | 64.57 | 6.945 | . | . | 3.1623 | . | -75.35 | -10.9767 |
| 7 | LOGIT | COV | 0 Converged | blast | . | . | . | . | . | . | . | -10.9767 |
| 8 | LOGIT | COV | 0 Converged | temp | -3483.23 | -223.669 | . | . | -75.3513 | . | 3808.42 | -10.9767 |
The data set pred created by the OUTPUT statement is displayed in Output 51.1.8. It contains all the variables in the input data set, the variable phat for the (cumulative) predicted probability, the variables lcl and ucl for the lower and upper confidence limits for the probability, and four other variables (IP_1, IP_0, XP_1, and XP_0) for the PREDPROBS= option. The data set also contains the variable _LEVEL_, indicating the response value to which phat, lcl, and ucl refer. For instance, for the first row of the OUTPUT data set, the values of _LEVEL_ and phat, lcl, and ucl are 1, 0.72265, 0.16892, and 0.97093, respectively; this means that the estimated probability that remiss=1 is 0.723 for the given explanatory variable values, and the corresponding 95% confidence interval is (0.16892, 0.97093). The variables IP_1 and IP_0 contain the predicted probabilities that remiss=1 and remiss=0, respectively. Note that values of phat and IP_1 are identical since they both contain the probabilities that remiss=1. The variables XP_1 and XP_0 contain the cross validated predicted probabilities that remiss=1 and remiss=0, respectively.
| Stepwise Regression on Cancer Remission Data |
| Predicted Probabilities and 95% Confidence Limits |
| Obs | remiss | cell | smear | infil | li | blast | temp | _FROM_ | _INTO_ | IP_0 | IP_1 | XP_0 | XP_1 | _LEVEL_ | phat | lcl | ucl |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 0.80 | 0.83 | 0.66 | 1.9 | 1.100 | 0.996 | 1 | 1 | 0.27735 | 0.72265 | 0.43873 | 0.56127 | 1 | 0.72265 | 0.16892 | 0.97093 |
| 2 | 1 | 0.90 | 0.36 | 0.32 | 1.4 | 0.740 | 0.992 | 1 | 1 | 0.42126 | 0.57874 | 0.47461 | 0.52539 | 1 | 0.57874 | 0.26788 | 0.83762 |
| 3 | 0 | 0.80 | 0.88 | 0.70 | 0.8 | 0.176 | 0.982 | 0 | 0 | 0.89540 | 0.10460 | 0.87060 | 0.12940 | 1 | 0.10460 | 0.00781 | 0.63419 |
| 4 | 0 | 1.00 | 0.87 | 0.87 | 0.7 | 1.053 | 0.986 | 0 | 0 | 0.71742 | 0.28258 | 0.67259 | 0.32741 | 1 | 0.28258 | 0.07498 | 0.65683 |
| 5 | 1 | 0.90 | 0.75 | 0.68 | 1.3 | 0.519 | 0.980 | 1 | 1 | 0.28582 | 0.71418 | 0.36901 | 0.63099 | 1 | 0.71418 | 0.25218 | 0.94876 |
| 6 | 0 | 1.00 | 0.65 | 0.65 | 0.6 | 0.519 | 0.982 | 0 | 0 | 0.72911 | 0.27089 | 0.67269 | 0.32731 | 1 | 0.27089 | 0.05852 | 0.68951 |
| 7 | 1 | 0.95 | 0.97 | 0.92 | 1.0 | 1.230 | 0.992 | 1 | 0 | 0.67844 | 0.32156 | 0.72923 | 0.27077 | 1 | 0.32156 | 0.13255 | 0.59516 |
| 8 | 0 | 0.95 | 0.87 | 0.83 | 1.9 | 1.354 | 1.020 | 0 | 1 | 0.39277 | 0.60723 | 0.09906 | 0.90094 | 1 | 0.60723 | 0.10572 | 0.95287 |
| 9 | 0 | 1.00 | 0.45 | 0.45 | 0.8 | 0.322 | 0.999 | 0 | 0 | 0.83368 | 0.16632 | 0.80864 | 0.19136 | 1 | 0.16632 | 0.03018 | 0.56123 |
| 10 | 0 | 0.95 | 0.36 | 0.34 | 0.5 | 0.000 | 1.038 | 0 | 0 | 0.99843 | 0.00157 | 0.99840 | 0.00160 | 1 | 0.00157 | 0.00000 | 0.68962 |
| 11 | 0 | 0.85 | 0.39 | 0.33 | 0.7 | 0.279 | 0.988 | 0 | 0 | 0.92715 | 0.07285 | 0.91723 | 0.08277 | 1 | 0.07285 | 0.00614 | 0.49982 |
| 12 | 0 | 0.70 | 0.76 | 0.53 | 1.2 | 0.146 | 0.982 | 0 | 0 | 0.82714 | 0.17286 | 0.63838 | 0.36162 | 1 | 0.17286 | 0.00637 | 0.87206 |
| 13 | 0 | 0.80 | 0.46 | 0.37 | 0.4 | 0.380 | 1.006 | 0 | 0 | 0.99654 | 0.00346 | 0.99644 | 0.00356 | 1 | 0.00346 | 0.00001 | 0.46530 |
| 14 | 0 | 0.20 | 0.39 | 0.08 | 0.8 | 0.114 | 0.990 | 0 | 0 | 0.99982 | 0.00018 | 0.99981 | 0.00019 | 1 | 0.00018 | 0.00000 | 0.96482 |
| 15 | 0 | 1.00 | 0.90 | 0.90 | 1.1 | 1.037 | 0.990 | 0 | 1 | 0.42878 | 0.57122 | 0.35354 | 0.64646 | 1 | 0.57122 | 0.25303 | 0.83973 |
| 16 | 1 | 1.00 | 0.84 | 0.84 | 1.9 | 2.064 | 1.020 | 1 | 1 | 0.28530 | 0.71470 | 0.47213 | 0.52787 | 1 | 0.71470 | 0.15362 | 0.97189 |
| 17 | 0 | 0.65 | 0.42 | 0.27 | 0.5 | 0.114 | 1.014 | 0 | 0 | 0.99938 | 0.00062 | 0.99937 | 0.00063 | 1 | 0.00062 | 0.00000 | 0.62665 |
| 18 | 0 | 1.00 | 0.75 | 0.75 | 1.0 | 1.322 | 1.004 | 0 | 0 | 0.77711 | 0.22289 | 0.73612 | 0.26388 | 1 | 0.22289 | 0.04483 | 0.63670 |
| 19 | 0 | 0.50 | 0.44 | 0.22 | 0.6 | 0.114 | 0.990 | 0 | 0 | 0.99846 | 0.00154 | 0.99842 | 0.00158 | 1 | 0.00154 | 0.00000 | 0.79644 |
| 20 | 1 | 1.00 | 0.63 | 0.63 | 1.1 | 1.072 | 0.986 | 1 | 1 | 0.35089 | 0.64911 | 0.42053 | 0.57947 | 1 | 0.64911 | 0.26305 | 0.90555 |
| 21 | 0 | 1.00 | 0.33 | 0.33 | 0.4 | 0.176 | 1.010 | 0 | 0 | 0.98307 | 0.01693 | 0.98170 | 0.01830 | 1 | 0.01693 | 0.00029 | 0.50475 |
| 22 | 0 | 0.90 | 0.93 | 0.84 | 0.6 | 1.591 | 1.020 | 0 | 0 | 0.99378 | 0.00622 | 0.99348 | 0.00652 | 1 | 0.00622 | 0.00003 | 0.56062 |
| 23 | 1 | 1.00 | 0.58 | 0.58 | 1.0 | 0.531 | 1.002 | 1 | 0 | 0.74739 | 0.25261 | 0.84423 | 0.15577 | 1 | 0.25261 | 0.06137 | 0.63597 |
| 24 | 0 | 0.95 | 0.32 | 0.30 | 1.6 | 0.886 | 0.988 | 0 | 1 | 0.12989 | 0.87011 | 0.03637 | 0.96363 | 1 | 0.87011 | 0.40910 | 0.98481 |
| 25 | 1 | 1.00 | 0.60 | 0.60 | 1.7 | 0.964 | 0.990 | 1 | 1 | 0.06868 | 0.93132 | 0.08017 | 0.91983 | 1 | 0.93132 | 0.44114 | 0.99573 |
| 26 | 1 | 1.00 | 0.69 | 0.69 | 0.9 | 0.398 | 0.986 | 1 | 0 | 0.53949 | 0.46051 | 0.62312 | 0.37688 | 1 | 0.46051 | 0.16612 | 0.78529 |
| 27 | 0 | 1.00 | 0.73 | 0.73 | 0.7 | 0.398 | 0.986 | 0 | 0 | 0.71742 | 0.28258 | 0.67259 | 0.32741 | 1 | 0.28258 | 0.07498 | 0.65683 |
Next, a different variable selection method is used to select prognostic factors for cancer remission, and an efficient algorithm is employed to eliminate insignificant variables from a model. The following statements invoke PROC LOGISTIC to perform the backward elimination analysis:
title 'Backward Elimination on Cancer Remission Data';
proc logistic data=Remission;
model remiss(event='1')=temp cell li smear blast
/ selection=backward fast slstay=0.2 ctable;
run;
The backward elimination analysis (SELECTION=BACKWARD) starts with a model that contains all explanatory variables given in the MODEL statement. By specifying the FAST option, PROC LOGISTIC eliminates insignificant variables without refitting the model repeatedly. This analysis uses a significance level of 0.2 to retain variables in the model (SLSTAY=0.2), which is different from the previous stepwise analysis where SLSTAY=.35. The CTABLE option is specified to produce classifications of input observations based on the final selected model.
Results of the fast elimination analysis are shown in Output 51.1.9 and Output 51.1.10. Initially, a full model containing all six risk factors is fit to the data (Output 51.1.9). In the next step (Output 51.1.10), PROC LOGISTIC removes blast, smear, cell, and temp from the model all at once. This leaves li and the intercept as the only variables in the final model. Note that in this analysis, only parameter estimates for the final model are displayed because the DETAILS option has not been specified.
| Backward Elimination on Cancer Remission Data |
| Model Information | ||
|---|---|---|
| Data Set | WORK.REMISSION | |
| Response Variable | remiss | Complete Remission |
| Number of Response Levels | 2 | |
| Model | binary logit | |
| Optimization Technique | Fisher's scoring | |
| Model Convergence Status |
|---|
| Convergence criterion (GCONV=1E-8) satisfied. |
Step 1. Fast Backward Elimination:
| Analysis of Effects Removed by Fast Backward Elimination | ||||||
|---|---|---|---|---|---|---|
| Effect Removed |
Chi-Square | DF | Pr > ChiSq | Residual Chi-Square |
DF | Pr > Residual ChiSq |
| blast | 0.0008 | 1 | 0.9768 | 0.0008 | 1 | 0.9768 |
| smear | 0.0951 | 1 | 0.7578 | 0.0959 | 2 | 0.9532 |
| cell | 1.5134 | 1 | 0.2186 | 1.6094 | 3 | 0.6573 |
| temp | 0.6535 | 1 | 0.4189 | 2.2628 | 4 | 0.6875 |
| Model Fit Statistics | ||
|---|---|---|
| Criterion | Intercept Only |
Intercept and Covariates |
| AIC | 36.372 | 30.073 |
| SC | 37.668 | 32.665 |
| -2 Log L | 34.372 | 26.073 |
| Testing Global Null Hypothesis: BETA=0 | |||
|---|---|---|---|
| Test | Chi-Square | DF | Pr > ChiSq |
| Likelihood Ratio | 8.2988 | 1 | 0.0040 |
| Score | 7.9311 | 1 | 0.0049 |
| Wald | 5.9594 | 1 | 0.0146 |
| Summary of Backward Elimination | |||||
|---|---|---|---|---|---|
| Step | Effect Removed |
DF | Number In |
Wald Chi-Square |
Pr > ChiSq |
| 1 | blast | 1 | 4 | 0.0008 | 0.9768 |
| 1 | smear | 1 | 3 | 0.0951 | 0.7578 |
| 1 | cell | 1 | 2 | 1.5134 | 0.2186 |
| 1 | temp | 1 | 1 | 0.6535 | 0.4189 |
Note that you can also use the FAST option when SELECTION=STEPWISE. However, the FAST option operates only on backward elimination steps. In this example, the stepwise process only adds variables, so the FAST option would not be useful.
Results of the CTABLE option are shown in Output 51.1.11.
| Classification Table | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Prob Level |
Correct | Incorrect | Percentages | ||||||
| Event | Non- Event |
Event | Non- Event |
Correct | Sensi- tivity |
Speci- ficity |
False POS |
False NEG |
|
| 0.060 | 9 | 0 | 18 | 0 | 33.3 | 100.0 | 0.0 | 66.7 | . |
| 0.080 | 9 | 2 | 16 | 0 | 40.7 | 100.0 | 11.1 | 64.0 | 0.0 |
| 0.100 | 9 | 4 | 14 | 0 | 48.1 | 100.0 | 22.2 | 60.9 | 0.0 |
| 0.120 | 9 | 4 | 14 | 0 | 48.1 | 100.0 | 22.2 | 60.9 | 0.0 |
| 0.140 | 9 | 7 | 11 | 0 | 59.3 | 100.0 | 38.9 | 55.0 | 0.0 |
| 0.160 | 9 | 10 | 8 | 0 | 70.4 | 100.0 | 55.6 | 47.1 | 0.0 |
| 0.180 | 9 | 10 | 8 | 0 | 70.4 | 100.0 | 55.6 | 47.1 | 0.0 |
| 0.200 | 8 | 13 | 5 | 1 | 77.8 | 88.9 | 72.2 | 38.5 | 7.1 |
| 0.220 | 8 | 13 | 5 | 1 | 77.8 | 88.9 | 72.2 | 38.5 | 7.1 |
| 0.240 | 8 | 13 | 5 | 1 | 77.8 | 88.9 | 72.2 | 38.5 | 7.1 |
| 0.260 | 6 | 13 | 5 | 3 | 70.4 | 66.7 | 72.2 | 45.5 | 18.8 |
| 0.280 | 6 | 13 | 5 | 3 | 70.4 | 66.7 | 72.2 | 45.5 | 18.8 |
| 0.300 | 6 | 13 | 5 | 3 | 70.4 | 66.7 | 72.2 | 45.5 | 18.8 |
| 0.320 | 6 | 14 | 4 | 3 | 74.1 | 66.7 | 77.8 | 40.0 | 17.6 |
| 0.340 | 5 | 14 | 4 | 4 | 70.4 | 55.6 | 77.8 | 44.4 | 22.2 |
| 0.360 | 5 | 14 | 4 | 4 | 70.4 | 55.6 | 77.8 | 44.4 | 22.2 |
| 0.380 | 5 | 15 | 3 | 4 | 74.1 | 55.6 | 83.3 | 37.5 | 21.1 |
| 0.400 | 5 | 15 | 3 | 4 | 74.1 | 55.6 | 83.3 | 37.5 | 21.1 |
| 0.420 | 5 | 15 | 3 | 4 | 74.1 | 55.6 | 83.3 | 37.5 | 21.1 |
| 0.440 | 5 | 15 | 3 | 4 | 74.1 | 55.6 | 83.3 | 37.5 | 21.1 |
| 0.460 | 4 | 16 | 2 | 5 | 74.1 | 44.4 | 88.9 | 33.3 | 23.8 |
| 0.480 | 4 | 16 | 2 | 5 | 74.1 | 44.4 | 88.9 | 33.3 | 23.8 |
| 0.500 | 4 | 16 | 2 | 5 | 74.1 | 44.4 | 88.9 | 33.3 | 23.8 |
| 0.520 | 4 | 16 | 2 | 5 | 74.1 | 44.4 | 88.9 | 33.3 | 23.8 |
| 0.540 | 3 | 16 | 2 | 6 | 70.4 | 33.3 | 88.9 | 40.0 | 27.3 |
| 0.560 | 3 | 16 | 2 | 6 | 70.4 | 33.3 | 88.9 | 40.0 | 27.3 |
| 0.580 | 3 | 16 | 2 | 6 | 70.4 | 33.3 | 88.9 | 40.0 | 27.3 |
| 0.600 | 3 | 16 | 2 | 6 | 70.4 | 33.3 | 88.9 | 40.0 | 27.3 |
| 0.620 | 3 | 16 | 2 | 6 | 70.4 | 33.3 | 88.9 | 40.0 | 27.3 |
| 0.640 | 3 | 16 | 2 | 6 | 70.4 | 33.3 | 88.9 | 40.0 | 27.3 |
| 0.660 | 3 | 16 | 2 | 6 | 70.4 | 33.3 | 88.9 | 40.0 | 27.3 |
| 0.680 | 3 | 16 | 2 | 6 | 70.4 | 33.3 | 88.9 | 40.0 | 27.3 |
| 0.700 | 3 | 16 | 2 | 6 | 70.4 | 33.3 | 88.9 | 40.0 | 27.3 |
| 0.720 | 2 | 16 | 2 | 7 | 66.7 | 22.2 | 88.9 | 50.0 | 30.4 |
| 0.740 | 2 | 16 | 2 | 7 | 66.7 | 22.2 | 88.9 | 50.0 | 30.4 |
| 0.760 | 2 | 16 | 2 | 7 | 66.7 | 22.2 | 88.9 | 50.0 | 30.4 |
| 0.780 | 2 | 16 | 2 | 7 | 66.7 | 22.2 | 88.9 | 50.0 | 30.4 |
| 0.800 | 2 | 17 | 1 | 7 | 70.4 | 22.2 | 94.4 | 33.3 | 29.2 |
| 0.820 | 2 | 17 | 1 | 7 | 70.4 | 22.2 | 94.4 | 33.3 | 29.2 |
| 0.840 | 0 | 17 | 1 | 9 | 63.0 | 0.0 | 94.4 | 100.0 | 34.6 |
| 0.860 | 0 | 17 | 1 | 9 | 63.0 | 0.0 | 94.4 | 100.0 | 34.6 |
| 0.880 | 0 | 17 | 1 | 9 | 63.0 | 0.0 | 94.4 | 100.0 | 34.6 |
| 0.900 | 0 | 17 | 1 | 9 | 63.0 | 0.0 | 94.4 | 100.0 | 34.6 |
| 0.920 | 0 | 17 | 1 | 9 | 63.0 | 0.0 | 94.4 | 100.0 | 34.6 |
| 0.940 | 0 | 17 | 1 | 9 | 63.0 | 0.0 | 94.4 | 100.0 | 34.6 |
| 0.960 | 0 | 18 | 0 | 9 | 66.7 | 0.0 | 100.0 | . | 33.3 |
Each row of the "Classification Table" corresponds to a cutpoint applied to the predicted probabilities, which is given in the Prob Level column. The  frequency tables of observed and predicted responses are given by the next four columns. For example, with a cutpoint of 0.5, 4 events and 16 nonevents were classified correctly. On the other hand, 2 nonevents were incorrectly classified as events and 5 events were incorrectly classified as nonevents. For this cutpoint, the correct classification rate is 20/27 (=74.1%), which is given in the sixth column. Accuracy of the classification is summarized by the sensitivity, specificity, and false positive and negative rates, which are displayed in the last four columns. You can control the number of cutpoints used, and their values, by using the PPROB= option.
frequency tables of observed and predicted responses are given by the next four columns. For example, with a cutpoint of 0.5, 4 events and 16 nonevents were classified correctly. On the other hand, 2 nonevents were incorrectly classified as events and 5 events were incorrectly classified as nonevents. For this cutpoint, the correct classification rate is 20/27 (=74.1%), which is given in the sixth column. Accuracy of the classification is summarized by the sensitivity, specificity, and false positive and negative rates, which are displayed in the last four columns. You can control the number of cutpoints used, and their values, by using the PPROB= option.
Copyright © SAS Institute, Inc. All Rights Reserved.