| The TREE Procedure |
| PROC TREE Statement |
The PROC TREE statement starts the TREE procedure.
The options that can appear in the PROC TREE statement are summarized in Table 91.1.
Option |
Description |
|---|---|
Data Sets |
|
DATA= |
specifies input data set |
DOCK= |
specifies that small clusters not be counted in OUT= data set |
LEVEL= |
defines disjoint cluster in OUT= data set |
NCLUSTERS= |
specifies number of clusters in OUT= data set |
OUT= |
specifies output data set |
ROOT= |
displays root of a subtree |
Cluster Heights |
|
HEIGHT= |
specifies variable for the height axis |
DISSIMILAR |
specifies that height values indicate dissimilarity |
SIMILAR |
specifies that height values indicate similarity |
Horizontal Trees |
|
HORIZONTAL |
specifies that height axis be horizontal |
Sort Order |
|
DESCENDING |
reverses sort order |
SORT |
sorts children by HEIGHT variable |
Displayed Output |
|
INC= |
specifies increment between tick values |
LINEPRINTER |
displays tree by using line printer graphics |
LIST |
displays all nodes in tree |
MAXHEIGHT= |
specifies maximum value on axis |
MINHEIGHT= |
specifies minimum value on axis |
NOPRINT |
suppresses display of tree |
NTICK= |
specifies number of tick intervals |
Graphics |
|
CFRAME= |
specifies color of the frame |
DESCRIPTION= |
specifies catalog description |
GOUT= |
specifies catalog name |
HAXIS= |
customizes horizontal axis |
HORDISPLAY= |
displays horizontal tree with leaves on right |
HPAGES= |
specifies number of pages to expand tree horizontally |
LINES= |
specifies line color and thickness, dots at nodes |
NAME= |
specifies name of graph in catalog |
VAXIS= |
customizes vertical axis |
VPAGES= |
specifies number of pages to expand tree vertically |
Line Printer Graphics |
|
PAGES= |
specifies number of pages |
POS= |
specifies number of column positions |
SPACES= |
specifies number of spaces between objects |
TICKPOS= |
specifies number of column positions between ticks |
FILLCHAR= |
specifies fill character between unjoined leaves |
JOINCHAR= |
specifies character displayed between joined leaves |
LEAFCHAR= |
specifies character representing clusters with no children |
TREECHAR= |
specifies character representing clusters with children |
- CFRAME=color
specifies a color for the frame, which is the rectangle bounded by the axes.
- DATA=SAS-data-set
specifies the input data set defining the tree. If you omit the DATA= option, the most recently created SAS data set is used.
- DESCENDING
- DES
- DESCRIPTION=entry-description
specifies a description for the graph in the GOUT= catalog. The default is "Proc Tree Graph Output."
- DISSIMILAR
- DIS
specifies that the values of the HEIGHT variable are dissimilarities; that is, a large height value means that the clusters are very dissimilar or far apart.
If neither the SIMILAR nor the DISSIMILAR option is specified, PROC TREE attempts to infer from the data whether the height values are similarities or dissimilarities. If PROC TREE cannot tell this from the data, it issues an error message and does not display a tree diagram.
- DOCK=n
causes observations in the OUT= data set assigned to output clusters with a frequency of
 or less to be given missing values for the output variables CLUSTER and CLUSNAME. If the NCLUSTERS= option is also specified, DOCK= also prevents clusters with a frequency of
or less to be given missing values for the output variables CLUSTER and CLUSNAME. If the NCLUSTERS= option is also specified, DOCK= also prevents clusters with a frequency of  or less from being counted toward the number of clusters requested by the NCLUSTERS= option. By default, DOCK=0.
or less from being counted toward the number of clusters requested by the NCLUSTERS= option. By default, DOCK=0. - FILLCHAR=’c’
- FC=’c’
specifies the character to display between leaves that are not joined into a cluster. The character should be enclosed in single quotes. The default is a blank. The LINEPRINTER option must also be specified.
- GOUT=<libref.>member-name
specifies the catalog in which the generated graph is stored. The default is WORK.GSEG.
- HAXIS=AXISn
specifies that the AXIS
 statement used to customize the appearance of the horizontal axis.
statement used to customize the appearance of the horizontal axis. - HEIGHT=name
- H=name
specifies certain conventional variables to be used for the height axis of the tree diagram. For many situations, the only option you need is the HEIGHT= option. Valid values for name and their meanings are as follows:
- HEIGHT | H
specifies the _HEIGHT_ variable.
- LENGTH | L
defines the height of each node as its path length from the root. This can also be interpreted as the number of ancestors of the node.
- MODE | M
specifies the _MODE_ variable.
- NCL | N
specifies the _NCL_ (number of clusters) variable.
- RSQ | R
specifies the _RSQ_ variable.
See also the section HEIGHT Statement, which can specify any variable in the input data set to be used for the height axis. In rare cases, you might need to specify either the DISSIMILAR option or the SIMILAR option.
- HORDISPLAY=RIGHT
specifies that the graph be oriented horizontally, with the leaf nodes on the right side, when the HORIZONTAL option is also specified. By default, the leaf nodes are on the left side.
- HORIZONTAL
- HOR
displays the tree diagram with the height axis oriented horizontally. The leaf nodes are on the side specified in the HORDISPLAY= option. If you do not specify the HORIZONTAL option, the height axis is vertical, with the root at the top. When the tree takes up more than one page, horizontal orientation can make the tree diagram considerably easier to read.
- HPAGES=n1
specifies that the original graph be enlarged to cover
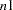 pages. If you also specify the VPAGES=
pages. If you also specify the VPAGES=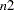 option, the original graph is enlarged to cover
option, the original graph is enlarged to cover 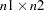 graphs. For example, if HPAGES=2 and VPAGES=3, then the original graph is generated, followed by
graphs. For example, if HPAGES=2 and VPAGES=3, then the original graph is generated, followed by 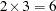 more graphs. In these six graphs, the original is enlarged by a factor of 2 in the horizontal direction and by a factor of 3 in the vertical direction. The graphs are generated in left-to-right and top-to-bottom order.
more graphs. In these six graphs, the original is enlarged by a factor of 2 in the horizontal direction and by a factor of 3 in the vertical direction. The graphs are generated in left-to-right and top-to-bottom order. - INC=n
specifies the increment between tick values on the height axis. If the HEIGHT variable is _NCL_, the default is usually 1, although a different value can be specified for consistency with other options. For any other HEIGHT variable, the default is some power of 10 times 1, 2, 2.5, or 5.
- JOINCHAR=’c’
- JC=’c’
specifies the character displayed between leaves that are joined into a cluster. The character should be enclosed in single quotes. The default is 'X'. The LINEPRINTER option must also be specified.
- LEAFCHAR=’c’
- LC=’c’
specifies the character used to represent clusters having no children. The character should be enclosed in single quotes. The default is a period. The LINEPRINTER option must also be specified.
- LEVEL=n
specifies the level of the tree defining disjoint clusters for the OUT= data set. The LEVEL= option also causes only clusters between the root and a height of
 to be displayed. The clusters in the output data set are those that exist at a height of
to be displayed. The clusters in the output data set are those that exist at a height of  on the tree diagram. For example, if the HEIGHT variable is _NCL_ (number of clusters) and LEVEL=5 is specified, then the OUT= data set contains five disjoint clusters. If the HEIGHT variable is _RSQ_ (
on the tree diagram. For example, if the HEIGHT variable is _NCL_ (number of clusters) and LEVEL=5 is specified, then the OUT= data set contains five disjoint clusters. If the HEIGHT variable is _RSQ_ (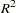 ) and LEVEL=0.9 is specified, then the OUT= data set contains the smallest number of clusters that yields an
) and LEVEL=0.9 is specified, then the OUT= data set contains the smallest number of clusters that yields an 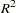 of at least 0.9.
of at least 0.9. - LINEPRINTER
specifies that the tree diagram be displayed using line printer graphics.
- LINES=( <COLOR=color> <WIDTH=n> <DOTS>)
specifies the color and the thickness of the lines of the tree, and whether a dot is drawn at each leaf node. Note that if the frame and the lines are specified to be the same color, PROC TREE selects a different color for the lines.
- LIST
lists all the nodes in the tree, displaying the height, parent, and children of each node.
- MAXHEIGHT=n
- MAXH=n
- MINHEIGHT=n
- MINH=n
- NAME=name
specifies the entry name for the generated graph in the GOUT= catalog. Note that each time another graph is generated with the same name, the name is modified by appending a number to make it unique.
- NCLUSTERS=n
- NCL=n
- N=n
specifies the number of clusters desired in the OUT= data set. The number of clusters obtained might not equal the number specified if (1) there are fewer than
 leaves in the tree, (2) there are more than
leaves in the tree, (2) there are more than  unconnected trees in the data set, (3) a multiway tree does not contain a level with the specified number of clusters, or (4) the DOCK= option eliminates too many clusters.
unconnected trees in the data set, (3) a multiway tree does not contain a level with the specified number of clusters, or (4) the DOCK= option eliminates too many clusters. The NCLUSTERS= option uses the _NCL_ variable to determine the order in which the clusters are formed. If there is no _NCL_ variable, the height variable (as determined by the HEIGHT statement or HEIGHT= option) is used instead.
- NTICK=n
specifies the number of tick intervals on the height axis. The default depends on the values of other options.
- NOPRINT
suppresses the display of the tree. Specify the NOPRINT option if you want only to create an OUT= data set.
- OUT=SAS-data-set
creates an output data set containing one observation for each object in the tree or subtree being processed and variables called CLUSTER and CLUSNAME showing cluster membership at any specified level in the tree. If you specify the OUT= option, you must also specify either the NCLUSTERS= or LEVEL= option in order to define the output partition level. If you want to create a permanent SAS data set, you must specify a two-level name (see "SAS Data Files" in SAS Language Reference: Concepts).
- PAGES=n
specifies the number of pages over which the tree diagram (from root to leaves) is to extend. The default is 1. The LINEPRINTER option must also be specified.
- POS=n
specifies the number of column positions on the height axis. The default depends on the value of the PAGES= option, the orientation of the tree diagram, and the values specified by the PAGESIZE= and LINESIZE= options. The LINEPRINTER option must also be specified.
- ROOT=’name’
specifies the value of the NAME variable for the root of a subtree to be displayed if you do not want to display the entire tree. If you also specify the OUT= option, the output data set contains only objects belonging to the subtree specified by the ROOT= option.
- SIMILAR
- SIM
specifies that the values of the HEIGHT variable represent similarities; that is, a large height value means that the clusters are very similar or close together.
If neither the SIMILAR nor the DISSIMILAR option is specified, PROC TREE attempts to infer from the data whether the height values are similarities or dissimilarities. If PROC TREE cannot tell this from the data, it issues an error message and does not display a tree diagram.
- SORT
sorts the children of each node by the HEIGHT variable, in the order of cluster formation. See the DESCENDING option on for details.
- SPACES=s
- S=s
specifies the number of spaces between objects in the output. The default depends on the number of objects, the orientation of the tree diagram, and the values specified by the PAGESIZE= and LINESIZE= options. The LINEPRINTER option must also be specified.
- TICKPOS=n
specifies the number of column positions per tick interval on the height axis. The default value is usually between 5 and 10, although a different value can be specified for consistency with other options.
- TREECHAR=’c’
- TC=’c’
specifies the character used to represent clusters with children. The character should be enclosed in single quotes. The default is 'X'. The LINEPRINTER option must also be specified.
- VAXIS=AXISn
specifies that the AXIS
 statement be used to customize the appearance of the vertical axis.
statement be used to customize the appearance of the vertical axis. - VPAGES=n2
specifies that the original graph be enlarged to cover
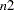 pages. If you also specify the HPAGES=
pages. If you also specify the HPAGES=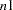 option, the original graph is enlarged to cover
option, the original graph is enlarged to cover 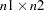 pages. For example, if HPAGES=2 and VPAGES=3, then the original graph is generated, followed by
pages. For example, if HPAGES=2 and VPAGES=3, then the original graph is generated, followed by 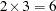 more graphs. In these six graphs, the original is enlarged by a factor of 2 in the horizontal direction and by a factor of 3 in the vertical direction. The graphs are generated in left-to-right and top-to-bottom order.
more graphs. In these six graphs, the original is enlarged by a factor of 2 in the horizontal direction and by a factor of 3 in the vertical direction. The graphs are generated in left-to-right and top-to-bottom order.
Copyright © 2009 by SAS Institute Inc., Cary, NC, USA. All rights reserved.