| The HPMIXED Procedure |
| PROC HPMIXED Statement |
The PROC HPMIXED statement invokes the procedure. You can specify the following options.
- DATA=SAS-data-set
names the SAS data set to be used by PROC HPMIXED. The default is the most recently created data set.
-
INFOCRIT=NONE
 |
| PQ
PQ |
| Q
Q
-
IC=NONE
 |
| PQ
PQ |
| Q
Q
determines the computation of information criteria in the "Fit Statistics" table. The criteria are all in smaller-is-better form, and are described in Table 43.1.
Table 43.1 Information Criteria Criteria
Formula
Reference
AIC
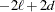
Akaike (1974)
AICC
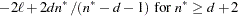
Hurvich and Tsai (1989) and
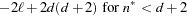
Burnham and Anderson (1998)
HQIC
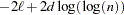
Hannan and Quinn (1979)
BIC
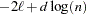
Schwarz (1978)
CAIC
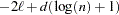
Bozdogan (1987)
Here
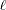 denotes the maximum value of the restricted log likelihood,
denotes the maximum value of the restricted log likelihood, 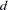 is the dimension of the model, and
is the dimension of the model, and  ,
, 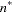 reflect the size of the data.
reflect the size of the data. The quantities
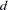 ,
,  , and
, and 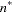 depend on the model and IC= option.
depend on the model and IC= option. models without random effects:
The IC=Q and IC=PQ options have no effect on the computation.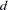 equals the number of parameters in the optimization whose solutions do not fall on the boundary or are otherwise constrained.
equals the number of parameters in the optimization whose solutions do not fall on the boundary or are otherwise constrained.  equals the number of used observations minus rank(X).
equals the number of used observations minus rank(X). 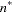 equals
equals  , unless
, unless 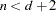 , in which case
, in which case 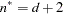 .
.
models with random effects:
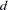 equals the number of parameters in the optimization whose solutions do not fall on the boundary or are otherwise constrained. If IC=PQ, this value is incremented by
equals the number of parameters in the optimization whose solutions do not fall on the boundary or are otherwise constrained. If IC=PQ, this value is incremented by 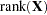 .
.  equals the effective number of subjects as displayed in the "Dimensions" table, unless this value equals 1, in which case
equals the effective number of subjects as displayed in the "Dimensions" table, unless this value equals 1, in which case  equals the number of levels of the first random effect specified. The IC=Q and IC=PQ options have no effect.
equals the number of levels of the first random effect specified. The IC=Q and IC=PQ options have no effect. 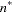 equals
equals  , unless
, unless 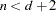 , in which case
, in which case 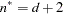 . The IC=Q and IC=PQ options have no effect.
. The IC=Q and IC=PQ options have no effect.
The IC=NONE option suppresses the "Fit Statistics" table. IC=Q is the default.
- ITDETAILS
displays the parameter values at each iteration and enables the writing of notes to the SAS log pertaining to "infinite likelihood" and "singularities" during optimization iterations.
- MAXCLPRINT=number
specifies the maximum levels of CLASS variables to print in the ODS table "ClassLevels." The default value is 20. MAXCLPRINT=0 enables you to print all levels of each CLASS variable. However, the option NOCLPRINT takes precedence over MAXCLPRINT.
- METHOD=REML
specifies the estimation method for the covariance parameters. The REML specification performs residual (restricted) maximum likelihood, and it is the default method. There is no other choice at this experimental stage.
- MMEQ
displays coefficients of the mixed model equations. These are
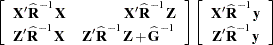
assuming
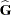 is nonsingular. If
is nonsingular. If 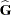 is singular, PROC HPMIXED produces the following coefficients
is singular, PROC HPMIXED produces the following coefficients 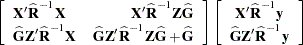
See the section "Model and Assumptions" for further information about these equations.
- NAMELEN=number
specifies the length to which long effect names are shortened. The default and minimum value is 20.
- NOCLPRINT<=number>
suppresses the display of the "Class Level Information" table if you do not specify number. If you do specify number, only levels with totals that are less than number are listed in the table.
- NOFIT
suppresses fitting of the model. When the NOFIT option is in effect, PROC HPMIXED produces the "Model Information," "Class Level Information," "Number of Observations," "Dimensions," and "Descriptive Statistics" tables. These can be helpful in gauging the computational effort required to fit the model.
- NOINFO
suppresses the display of the "Model Information," "Number of Observations," and "Dimensions" tables.
- NOITPRINT
- NOPRINT
suppresses the normal display of results. The NOPRINT option is useful when you want only to create one or more output data sets with the procedure by using the OUTPUT statement. Note that this option temporarily disables the Output Delivery System (ODS); see Chapter 20, Using the Output Delivery System, for more information.
- NOPROFILE
includes the residual variance as one of the covariance parameters in the optimization iterations. This option applies only to models that have a residual variance parameter. By default, this parameter is profiled out of the optimization iterations, except when you have specified the HOLD= option in the PARMS statement.
-
ORDER=DATA
 |
| FORMATTED
FORMATTED |
| FREQ
FREQ |
| INTERNAL
INTERNAL
specifies the sorting order for the levels of all CLASS variables. This ordering determines which parameters in the model correspond to each level in the data, so the ORDER= option can be useful when you use CONTRAST or ESTIMATE statements.
The default is ORDER=FORMATTED.
The following table shows how PROC HPMIXED interprets values of the ORDER= option.
Value of ORDER=
Levels Sorted By
DATA
order of appearance in the input data set
FORMATTED
external formatted value, except for numeric
variables with no explicit format, which are
sorted by their unformatted (internal) value
FREQ
descending frequency count; levels with the
most observations come first in the order
INTERNAL
unformatted value
For FORMATTED and INTERNAL, the sort order is machine dependent. For more information about sorting order, see the chapter about the SORT procedure in the Base SAS Procedures Guide and the discussion of BY-group processing in SAS Language Reference: Concepts.
- SINGCHOL=number
tunes the singularity criterion in Cholesky decompositions. The default is 1E6 times the machine epsilon; this product is approximately 1E
 10 on most computers.
10 on most computers. - SINGRES=number
sets the tolerance for which the residual variance is considered to be zero. The default is 1E4 times the machine epsilon; this product is approximately 1E
 12 on most computers.
12 on most computers. - SINGULAR=number
tunes the general singularity criterion applied by the HPMIXED procedure in divisions and inversions. The default is 1E4 times the machine epsilon; this product is approximately 1E
 12 on most computers.
12 on most computers.
Note: This procedure is experimental.
Copyright © 2009 by SAS Institute Inc., Cary, NC, USA. All rights reserved.