| The GLIMMIX Procedure |
G-Side and R-Side Random Effects and Covariance Structures
The GLIMMIX procedure distinguishes two types of random effects. Depending on whether the parameters of the covariance structure for random components in your model are contained in 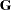 or in
or in 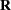 , the procedure distinguishes between "G-side" and "R-side" random effects. The associated covariance structures of
, the procedure distinguishes between "G-side" and "R-side" random effects. The associated covariance structures of 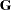 and
and 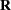 are similarly termed the G-side and R-side covariance structure, respectively. R-side effects are also called "residual" effects. Simply put, if a random effect is an element of
are similarly termed the G-side and R-side covariance structure, respectively. R-side effects are also called "residual" effects. Simply put, if a random effect is an element of 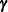 , it is a G-side effect and you are modeling the G-side covariance structure; otherwise, you are modeling the R-side covariance structure of the model. Models without G-side effects are also known as marginal (or population-averaged) models. Models fit with the GLIMMIX procedure can have none, one, or more of each type of effect.
, it is a G-side effect and you are modeling the G-side covariance structure; otherwise, you are modeling the R-side covariance structure of the model. Models without G-side effects are also known as marginal (or population-averaged) models. Models fit with the GLIMMIX procedure can have none, one, or more of each type of effect.
Note that an R-side effect in the GLIMMIX procedure is equivalent to a REPEATED effect in the MIXED procedure. The R-side covariance structure in the GLIMMIX procedure is the covariance structure that you would formulate with the REPEATED statement in the MIXED procedure. In the GLIMMIX procedure all random effects and their covariance structures are specified through the RANDOM statement. See the section Comparing the GLIMMIX and MIXED Procedures for a comparison of the GLIMMIX and MIXED procedures.
The columns of 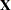 are constructed from effects listed on the right side in the MODEL statement. Columns of
are constructed from effects listed on the right side in the MODEL statement. Columns of 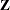 and the variance matrices
and the variance matrices 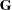 and
and 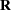 are constructed from the RANDOM statement.
are constructed from the RANDOM statement.
The 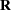 matrix is by default the scaled identity matrix,
matrix is by default the scaled identity matrix, 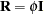 . The scale parameter
. The scale parameter 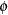 is set to one if the distribution does not have a scale parameter, such as in the case of the binary, binomial, Poisson, and exponential distribution (see Table 38.15). To specify a different
is set to one if the distribution does not have a scale parameter, such as in the case of the binary, binomial, Poisson, and exponential distribution (see Table 38.15). To specify a different 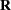 matrix, use the RANDOM statement with the _RESIDUAL_ keyword or the RESIDUAL option. For example, to specify that the Time effect for each patient is an R-side effect with a first-order autoregressive covariance structure, use the RESIDUAL option:
matrix, use the RANDOM statement with the _RESIDUAL_ keyword or the RESIDUAL option. For example, to specify that the Time effect for each patient is an R-side effect with a first-order autoregressive covariance structure, use the RESIDUAL option:
random time / type=ar(1) subject=patient residual;
To add a multiplicative overdispersion parameter, use the _RESIDUAL_ keyword:
random _residual_;
You specify the link function 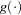 with the LINK= option in the MODEL statement or with programming statements. You specify the variance function that controls the matrix
with the LINK= option in the MODEL statement or with programming statements. You specify the variance function that controls the matrix  with the DIST= option in the MODEL statement or with programming statements.
with the DIST= option in the MODEL statement or with programming statements.
Unknown quantities subject to estimation are the fixed-effects parameter vector 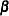 and the covariance parameter vector
and the covariance parameter vector 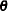 that comprises all unknowns in
that comprises all unknowns in 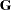 and
and 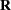 . The random effects
. The random effects 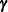 are not parameters of the model in the sense that they are not estimated. The vector
are not parameters of the model in the sense that they are not estimated. The vector 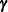 is a vector of random variables. The solutions for
is a vector of random variables. The solutions for 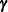 are predictors of these random variables.
are predictors of these random variables.
Copyright © 2009 by SAS Institute Inc., Cary, NC, USA. All rights reserved.